Institute of Oceanology, Chinese Academy of Sciences
Article Information
- WANG Zhaohui, LEI Mingdan, JI Shuanghui, XIE Changliang, CHEN Jiazhuo, LI Weiguo, JIANG Tao
- Comparison in diversity of eukaryotic algae in surface sediments from different functional sea areas of Qingdao coast, the Yellow Sea, China: a metabarcoding approach
- Journal of Oceanology and Limnology, 40(6): 2322-2342
- http://dx.doi.org/10.1007/s00343-021-1200-0
Article History
- Received Jun. 22, 2021
- accepted in principle Aug. 11, 2021
- accepted for publication Oct. 8, 2021
2 Engineering Research Center of Tropical and Subtropical Aquatic Ecological Engineering, Ministry of Education, Guangzhou 510632, China;
3 School of Ocean, Yantai University, Yantai 264005, China
Marine sediments provide luxuriant nutrients for benthic organisms, and are considered as "seed banks" of biodiversity in the seawater, out of which species can reemerge into the water column in appropriate opportunity (Massana et al., 2015). As the main producers of benthic eukaryotic communities, eukaryotic algae are major components in microbial food webs, playing important roles in energy flow and biogeochemical circle in the benthic ecosystems (Bik et al., 2012). Benthic microalgae have been regarded as an important ecological indicator for assessing anthropogenic effects on the marine environments (Blanco et al., 2008; Fierro et al., 2019).
Many phytoplankton species have the ability to experience the dormant phase as a part of their life circle under the stresses of adverse environmental conditions in the forms of resting spores and resting cysts (hereafter referred to as resting stages) (Head, 1996). The resting stages thicken the cell wall and sink to the sea floor, and then become a part of benthic community (Piredda et al., 2017). The resting stages have thick resistant wall, and can withstand low temperature, high pressure, hypoxic and dark benthic environment, and can survive in sediments for hundreds of years (reviewed by Ellegaard and Ribeiro, 2018). When the environmental conditions are suitable, the resting stages germinate and release vegetative cells into the water column (McQuoid, 2002). Phytoplankton resting stages are regarded as the "seed banks" of algal blooms, and play important role in initiating and dissipation of harmful algal blooms (HABs) (Smayda, 2002; Genovesi et al., 2009).
Identification of benthic microalgae has traditionally been based on microscopic observations and cell counting after separating cells from sediments. However, this approach has significant limitations, such as the recovery rate during separation and concentration, biases in the recovered groups, and difficulties associated with morphological identification (Piredda et al., 2017). These limitations have been overcome with the implementation of modern molecular based metabarcoding techniques, particular for the utilization of high-throughput sequencing (HTS) techniques (Penna et al., 2017).
Metabarcoding allows identification of microorganisms without taxonomic expertise, and only need to match the HTS-derived DNA fragments to the reference sequence libraries (Keeley et al., 2018). Standardized protocols can be developed and the results are auditable (Ji et al., 2013). HTS based metabarcoding has become a powerful and rapid technique to identify and quantify benthic organisms for biomonitoring purposes (Keeley et al., 2018).
The coastal zone is the most active area of human activity, and provides abundant resources and have high ecological value and environmental functions. However, many coastal environments have been suffering from problems of environmental degradation, decline in biodiversity, and species invasion as the rapid economic development, population expansion, and over-exploitation of marine resources (Yu et al., 2019). Qingdao is located in the eastern China, southeast of Shandong Peninsula and east of the Yellow Sea. It is the economic center of Shandong Province as the important international port, the modern marine industry zone, the international shipping hub in Northeast Asia, and the maritime sports base. Qingdao City covered 758.16 km2 area with the population of 9.50 million in 2019. Due to the rapid increase of economic development and population size as well as the increase of aquaculture, the pollution level in Qingdao coastal waters has been increasing recently (Yuan et al., 2017). The degradation of water quality results in the frequent occurrence of algal blooms. A total of 36 algal blooms occurred in the coastal areas of Qingdao between 1990 and 2017, in which more than 80% occurred in Jiaozhou Bay (Zhou et al., 2020). Moreover, sediments in Jiaozhou Bay have been polluted to a certain extent (Gu et al., 2021; Wang et al., 2021).
Intensive surveys have been carried out to investigate phytoplankton community in Jiaozhou Bay during the past fifty years, and results have shown rich phytoplankton diversity and great variations of the dominant phytoplankton taxa with time (reviewed by Liu and Chen, 2021). Meanwhile, metabarcoding studies revealed much more phytoplankton species in Jiaozhou Bay than the routine microscopic observations (Liu et al., 2020b; Chen et al., 2021). However, there are few studies on phytoplankton resting stages in sediments, with only one report on dinoflagellate cysts in Jiaozhou Bay based on microscopic analysis (Li et al., 2017), which cannot fully reflect the diversity and distribution of phytoplankton resting stages in sediments. In order to understand the distribution of phytoplankton resting stages and the potential of HABs in the Qingdao coast, surface sediments were collected from three different function sea areas of the Qingdao coast in this study, including a eutrophic enclosed bay (the inner Jiaozhou Bay), a conservation area (the Qingdao Amphioxus Reserve area), and a scenic and aquacultural area (the Laoshan Coast). HTS-based metabarcoding was used to investigate diversity and community structure of eukaryotic algae. Biogenic elements including total organic carbon (TOC), organic matter (OM), total nitrogen (TN), total phosphorus (TP), and biogenic silicon (BSi) were analyzed, and the relationships between algal community and biogenic elements were discussed. The purpose of this study was to compare benthic microalgae composition among the three sea areas with different degrees of human activities, and to discuss the distribution of resting stages of HAB species in sediments from the Qingdao coast. This study represents a first attempt to study eukaryotic algae in sediments from the Qingdao coast using the metabarcoding approach.
2 MATERIAL AND METHOD 2.1 Study areas and sediment collectionQingdao is located in the southeast of the Shandong Peninsula, which borders the Yellow Sea in the east and the south. Surface sediments were collected in three sea areas along the Qingdao coast, i.e., the inner Jiaozhou Bay, the Laoshan Coast, and the Qingdao Amphioxus Reserve area (Fig. 1). Jiaozhou Bay, in the northwest of Qingdao city center, is a large semi-closed bay connected to the Yellow Sea by a narrow bay mouth. Jiaozhou Bay has multiple functions, such as port, transportation, and aquaculture. The Laoshan Coast locates in the eastern part of Qingdao City and surround the famous tourist attraction the Laoshan Scenic Area. There are many aquaculture farms in the Laoshan Coast. The Amphioxus Reserve area is located in the southern part of Qingdao City, just outside the mouth of the Jiaozhou Bay and close to the Qingdao port. The Amphioxus Reserve area was established in August 2004 to protect wild amphioxus population and its habitat environment.
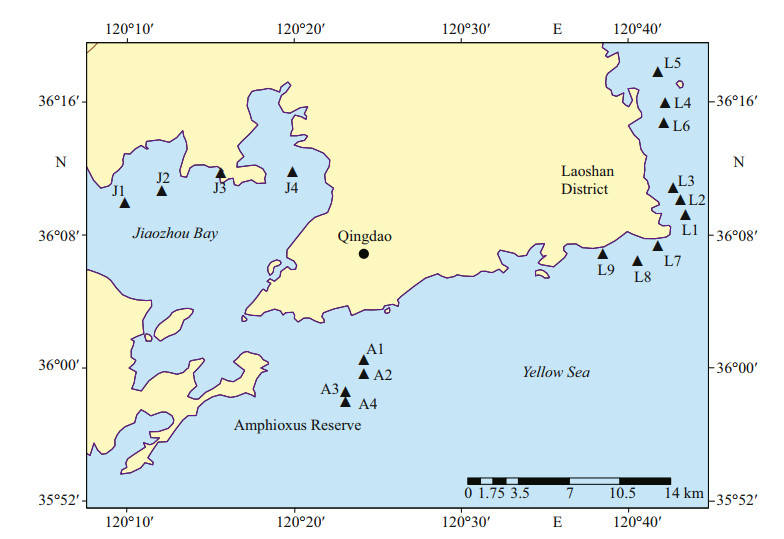
|
| Fig.1 Sampling stations in Qingdao coastal sea area Stations J1–J4 are in the inner Jiaozhou Bay, stations A1–A4 in the Amphioxus Reserve area, and stations L1–L9 in the Laoshan Coast. |
Surface sediments were collected in triplicate from seventeen stations in the three sea areas using a Peterson grab between November and December 2017 (Fig. 1). The top 2 cm of sediments were sampled with a polyethylene spatula, and the three replicate samples were mixed well together as one sample and sealed in a polyethylene bag, and then stored in the cold (4 ℃) dark condition for six months before DNA extraction to inactivate most of the microalgal vegetative cells, and to make sure that most of the survives were resting stages (Piredda et al., 2017).
2.2 DNA extraction, PCR amplification, and sequencingGenomic DNA of ca. 1.0 g of sediment sample was extracted using DNeasy PowerSoil Kit (Qiagen, Germany) according to the manufacturer's instructions. Polymerase chain reaction (PCR) amplification of ca. 450-bp fragment of the hypervariable V4 region of the 18S rRNA gene (rDNA) was carried out with the universal eukaryotic primers 3NDf 5′-GGCAAGTCTGGTGCCAG-3′ (Cavalier-Smith et al., 2009) and V4_euk_R2 5′-ACGGTATCTATCTCTTCG-3′ (Bråte et al., 2010). PCR products were then sequenced on an Illumina MisSeq PE300 platform (Illumina, San Diego, CA). Sequencing was performed by Majorbio Bio-pharm Technology Co., Ltd. (Shanghai, China).
2.3 Sequence data cleaning, filtering, and OTU analysisRaw data from the Illumina libraries were assigned to individual samples by their barcodes. Paired-end reads were truncated to remove unique barcode and primer sequences, then assembled using FLASH (Version 1.2.7). The obtained filtered sequences witherror were removed via using QIIME (QIIME, version 1.8, http://qiime.org) software. The Operational Taxonomic Unit (OTU) was the sequence similarity threshold, and sequences with similarities above the threshold were merged into the same OTU at 97% sequence similarity by using UPARSE (version 7.1, http://drive5.com/uparse/). Singletons and doubletons were removed from the dataset, and the chimeras were removed using USEARCH (version 8.1.1861, Http://www.drive5.com/Usearch/). The taxonomic assignment of each OTU was classified against the PR2 database (version 4.12.0, https://pr2-database.org/, searched on 27 March 2021) using the Ribosomal Database Project (RDP) classifier (Caporaso et al., 2010) with a bootstrap confidence of 80%. Those OTUs, which were not assigned to the species level in PR2 database, were further annotated against the NCBI database (https://www.ncbi.nlm.nih.gov/, searched on 17 April 2021) with the highest similarity (97%). The raw data were deposited into the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) (https://www.ncbi.nlm.nih.gov/sra/) with the accession number PRJNA722324. The classification of algal taxa is based on taxonomic information in AlgaeBase, a global algal database (Guiry and Guiry, 2021, https://www.algaebase.org/). A taxon was annotated as an HAB species if it is categorized as harmful microalgae in the online version of the Intergovernmental Oceanographic Commission (IOC) Taxonomic Reference List of Harmful Micro Algae (Lundholm et al., 2009 onwards) or in "Manual on Harmful Marine Microalgae" (Hallegraeff et al., 2003) or had been reported as an HAB species in previous studies.
2.4 Analyses of biogenic elementsSediments for biogenic elements analysis were dried in an oven at 40 ℃ until a constant weight was reached. The dried sediments were ground gently with an agate mortar and pestle, sieved through a 100-μm mesh for homogenization and stored in sealed plastic bags. The biogenic elements were measured in triplicate. TOC and TN was measured by a Perkin-Elmer 2400 Series II CHNS/O Analyzer (Perkin Elmer Inc., USA). TP was measured by potassium persulfate digestion method (Thien and Myers, 1992). OM was determined by ignition loss in a muffle furnace (SG-XL1200, Honglang, Shanghai, China). BSi was measured by the molybdate blue spectrophotometric method after removing the carbonates and organics by 1-mol/L HCl and 10% H2O2 and digested using 0.5-mol/L Na2CO3 solution (Mortlock and Froelich, 1989). The quality assurance/quality control (QA/QC) was assessed by the analyses of blank reagents and five replicates of the certified reference material (Offshore Marine Sediment, GBW 07314). The analytical precision was controlled to within 5% for biogenic elements.
2.5 Statistical analysesRarefaction curves were generated to assess the degree of sample saturation using the R 4.0.0 package vegan. The alpha diversity of eukaryotic organisms and eukaryotic algae, including OTU richness, Margalef, Simpson diversity, Shannon diversity, Pielou index, Chao1 and ACE, were analyzed after rarefying each sample to the number of DNA reads in the sample with the fewest reads (35 984 reads in this study). The Bray-Curtis (BC) index was used as a measure of similarity between the samples (beta-diversity). A distance matrix was computed with the BC index, and a hierarchical cluster tree was constructed using the un-weighted pair group method with arithmetic mean (UPGMA) with bootstrap support. Venn diagrams were calculated and plotted based on different samples and sea areas. All these analyses were conducted using the R 4.0.0 package vegan. A phylogenetic tree was constructed to illustrate the relationships between the algal communities in surface sediments from the Qingdao coast using the Neighbor-Joining algorithm method with the maximum likelihood model (MEGA v. 10.0 software) with bootstrap values of 1 000 replicate runs. A bubble chart of the top 25 abundance OTUs was drawn with the R package ggplot2, reshape2, and formats according to their abundances, and the DNA reads were logarithmically transformed. The redundancy analysis (RDA) was performed to revel the impacts of biogenic elements in explaining the distribution patterns of algal communities in seventeen surface sediments using Canoco4.5. All data were logarithmically transformed to obtain the equal weight of the elements in RDA analysis.
3 RESULT 3.1 Sequence dataA total of 947 886 eukaryotic DNA sequences and 1 497 OTUs were obtained in 17 surface sediment samples from the Qingdao coast. Rarefaction curves suggested adequate coverage for all datasets (Fig. 2). DNA reads in each sample ranged from 35 984 at station L1 to 72 247 at station L3, and OTU richness were between 175 and 684 OTUs, with the lowest at station J3 and the highest at station L2, respectively (Figs. 2–3). There were 171 722 algal sequences and 207 algal OTUs. The number of algal sequences at each station varied greatly (Fig. 3a), ranging from 424 to 28 082 reads with the highest at station L6 and the lowest at station J3. The proportions of algal reads to the total eukaryotic reads ranged from 0.59% at station J3 to 41.25% at station L6. The algal OTU richness of each sample ranged from 20 OTUs at station J3 to 103 OTUs at station L6 with the proportions of 8.93%–20.80% (Fig. 3b).
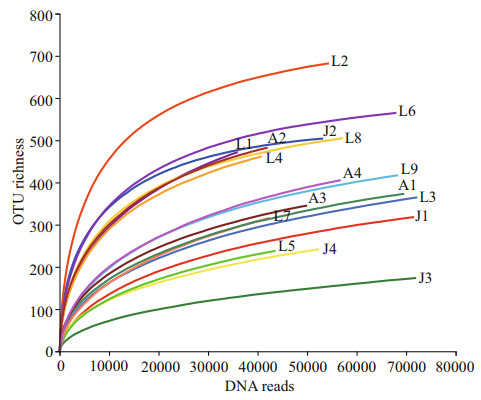
|
| Fig.2 Rarefaction curves of DNA reads based on OTUs at 97% sequence similarity |
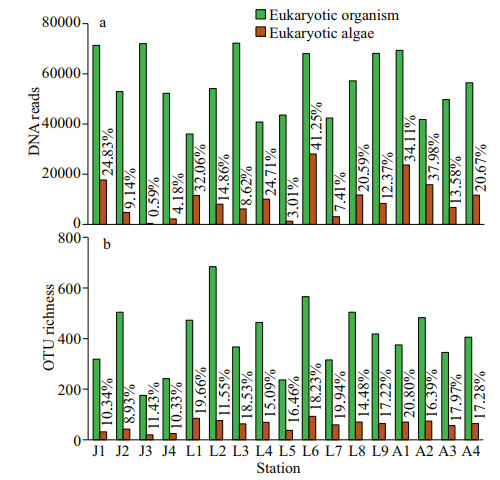
|
| Fig.3 DNA reads (a) and OTU richness (b) of eukaryotic organisms and eukaryotic algae in surface sediments from 17 sites of the Qingdao coastal |
The eukaryotic algae detected in this study included 75 genera in 24 classes of 8 phyla, i.e., Bacillariophyceae and Mediophyceae in Bacillariophyta, Chlorophyceae, Chlorodendrophyceae, Chloropicophyceae, Mamiellophyceae, Palmophyllophyceae, Pedinophyceae, Pyramimonadophyceae, Trebouxiophyceae, and Ulvophyceae in Chlorophyta, Cryptophyceae in Cryptophyta, Dinophyceae, and Syndiniophyceae in Dinophyta, Prymnesiophyceae in Haptophyta, Chrysophyceae, Eustigmatophyceae, Pelagophyceae, Phaeophyceae, Raphidophyceae, and Xanthophyceae in Ochrophyta, Prasinodermatophyceae in Prasinodermatophyta, and Florideophyceae and Rhodellophyceae in Rhodophyta. There were 124 OTUs (59.9%) identified at the species level after BLAST against the PR2 and the NCBI databases, and 161 OTUs (77.8%) at the genus level, and other 46 OTUs (22.2%) only into class level. Sometimes more than one or two OTUs were annotated as the same taxa, and altogether 98 species were detected in this study, including 12 Bacillariophyta (13 OTUs), 23 Chlorophyta (27 OTUs), 2 Cryptophyta (2 OTUs), 45 Dinophyta (66 OTUs), 1 Haptophyta (1 OTU), 13 Ochrophyta (13 OTUs), and 2 Rhodophyta (3 OTUs) species (Supplementary Table S1). There were 47, 89, and 61 species recorded in samples from the inner Jiaozhou Bay, the Laoshan Coast, and the Amphioxus Reserve area, respectively. Most of the species or genera identified in surface sediments in this study were marine species, some were brackish or marine/freshwater species, and terrestrial and benthic species were also detected. About 47 taxa have been reported to form resting stages, and 12 species are firstly recorded in the Chinese coastal waters (Supplementary Table S1).
Based on DNA reads at the class level (Fig. 4a), Dinophyceae dominated in the eukaryotic algal assemblages in surface sediments from the Qingdao coast, with the total of 124 516 reads accounting for 72.51% of total algal sequences. The second dominant class was Chrysophyceae, with 41 137 reads and proportion of 23.96%. Diatoms, including two classes, Mediophyceae and Bacillariophyceae, in this study, ranked the third with 2 197 reads and proportion of 1.27%. There were 1 178 reads of raphidophytes with the proportion of 0.69%. Syndiniophytes contributed 0.51% to the total algal reads. Sequences of other classes were less than 500 reads and contributed low than 0.5% of the algal reads.
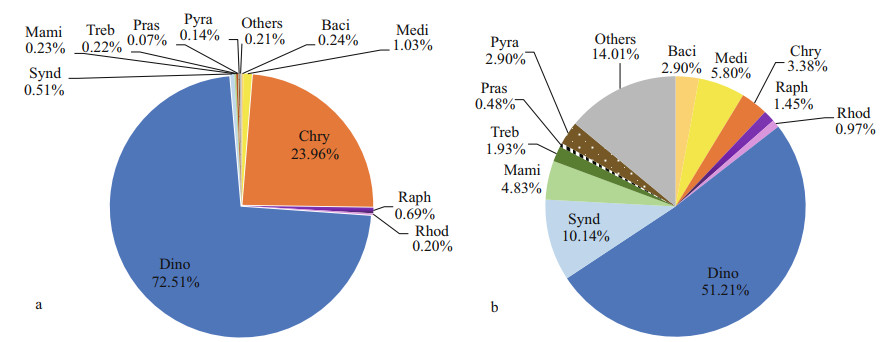
|
| Fig.4 Overall relative abundances of DNA reads (a) and OTU richness (b) of eukaryotic algae at the class level Classes are represented by their first four characters. Baci: Bacillariophyceae; Medi: Mediophyceae; Chry: Chrysophyceae; Raph: Raphidophyceae; Rhod: Rhodellophyceae; Dino: Dinophyceae; Synd: Syndiniophyceae; Mami: Mamiellophyceae; Treb: Trebouxiophyceae; Pras: Prasinodermatophyceae; Pyra: Pyramimonadophyceae; others: Chlorodendrophyceae, Chlorophyceae, Chloropicophyceae, Cryptophyceae, Eustigmatophyceae, Florideophyceae, Palmophyllophyceae, Pedinophyceae, Pelagophyceae, Phaeophyceae, Prymnesiophyceae, Ulvophyceae, and Xanthophyceae. |
Dinoflagellates (Dinophyta, including Dinophyceae and Syndiniophyceae) were the most diverse algal group in this study, with 106 OTUs and 21 OTUs, respectively, together which represented 61.35% of the total algae OTUs (Fig. 4b). However, none of the syndiniophytes were identified at the species level. Thirty-three OTUs (15.94%) were recorded in nine classes of Chlorophyta. Among them, 10 OTUs (4.83%) were recorded in the marine green algal class Mamiellophyceae (Fig. 4b). Diatoms were the third diverse group, with 12 OTUs (5.80%) and 6 OTUs (2.90%) for Mediophyceae and Bacillariophyceae, respectively.
To further understand the relationships among eukaryotic algae, a phylogenetic tree was constructed based on all algal OTUs detected in this study (Fig. 5). All OTUs in Dinophyta cluster into a big group, while OTUs in the other seven phyla form up another big group. The second group is separated into two large clades: one including Chlorophyta and the other three phyla (Rhodophyta, Cryptophyta, and Haptophyta). However, Ochrophyta and Bacillariophyta cluster to form another clade. OTUs in Chlorophyta join together as a large sub-clade, and those in Rhodophyta, Cryptophyta, and Haptophyta cluster. Generally, OTUs of the same phylum clustered. However, an unclassified diatom OTU (OTU241) and three unclassified Ochrophyta OTUs cluster into a small branch in the diatom clade. In addition, an OTU of Prasinodermatophyta, a new algal phylum which was recently separated from Chlorophyta (Li et al., 2020), is within the large branch of Chlorophyta.
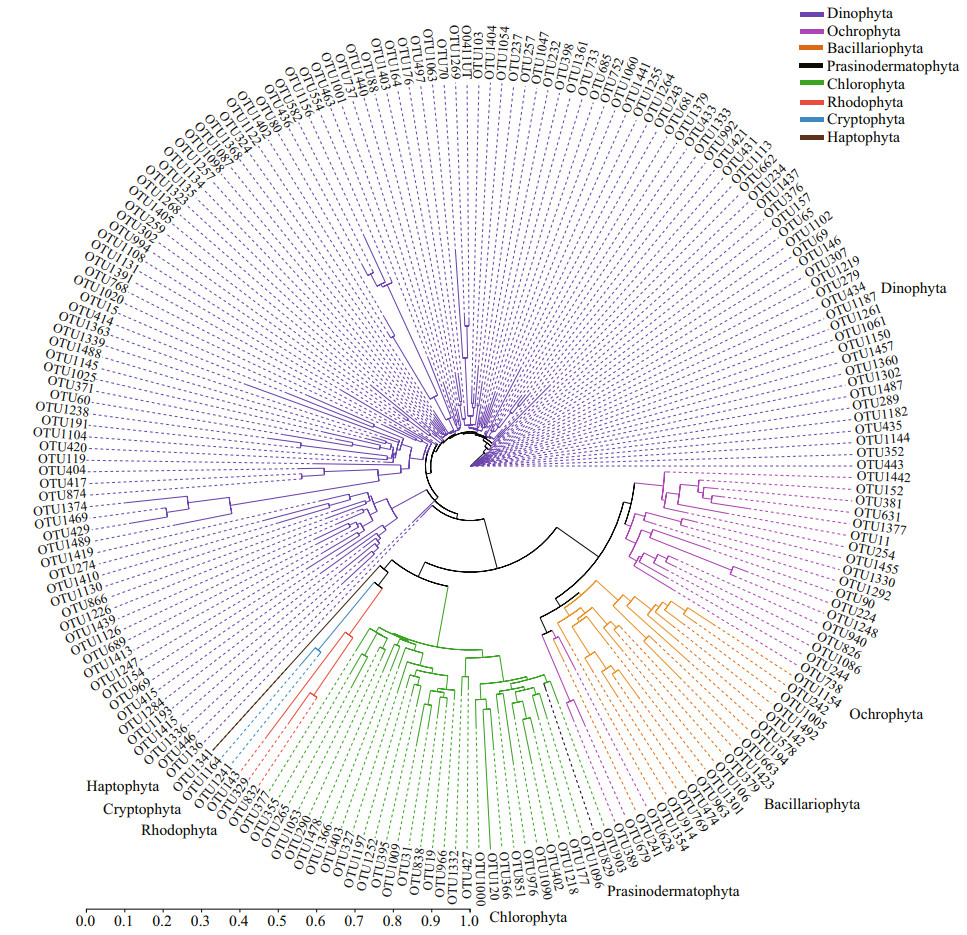
|
| Fig.5 A phylogenetic tree on OTU lever based on eukaryotic algal sequences in surface sediments from the Qingdao coast using the Neighbor-Joining algorithm method with the maximum likelihood model (MEGA v.10.0) |
The composition of algal community was quite different in the three sea areas of the Qingdao coast (Fig. 6). The eukaryotic algal community in the inner Jiaozhou Bay (JZ) was dominated by chrysophytes (Fig. 6a), which contributed to 77.40% of the total algal sequences; followed by dinoflagellates (including Dinophyceae and Syndiniophyceae) accounting for 17.79% of the algal sequences. However, dinoflagellates predominated in the Laoshan Coast (LS) with a relative percentage of 95.25%. Dinoflagellates and chrysophytes co-dominated in the Amphioxus Reserve area (AR) with the percentages of 62.98% and 32.28%, respectively. The compositions of OTU richness were comparable in the three sea areas (Fig. 6b), in which higher diversities were observed in dinoflagellates (Dinophyceae and Syndiniophyceae), diatoms (Mediophyceae and Bacillariophyceae), and Mamiellophyceae of Chlorophyta. Dinoflagellates accounted for 47.76%, 63.64%, and 66.12% of the algal OTUs in JZ, LS, and AR, respectively.
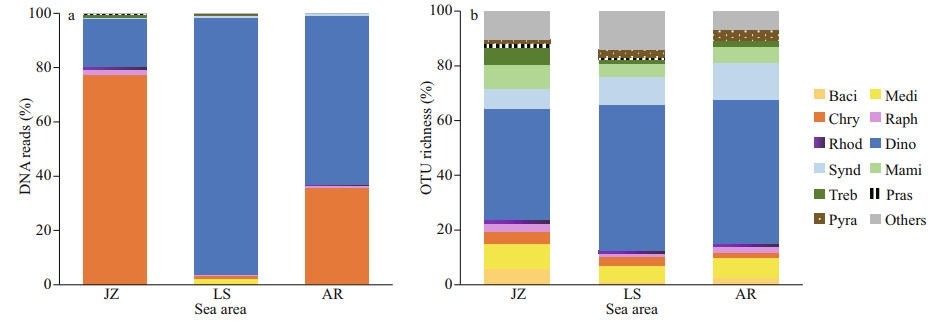
|
| Fig.6 Relative abundance of DNA reads (a) and OTU richness (b) of eukaryotic algae in surface sediments from the three sea areas of the Qingdao coast JZ: Jiaozhou Bay; LS: the Laoshan Coast; AR: the Amphioxus Reserve area. Classes are represented by their first four characters the same as shown in Fig. 4. |
The Venn diagrams highlighted the differences of algal communities among the seventeen samples in the three sea areas (Fig. 7). Only 44 OTUs (21.3%) shared among the three sea areas, and 2 OTUs shared among all samples. Particularly, the Laoshan Coast (LS) had the highest number of OTUs, and 66 unique OTUs (31.9%) were detected in this sea area (Fig. 7a). The numbers of unique OTUs were comparable in the inner Jiaozhou Bay (JZ) and the Amphioxus Reserve area (AR), which accounted for 4.3% of the total OTUs (Fig. 7a). The reason for the high OTU richness in the Laoshan Coast was partly due to more samples collected (9 samples), while only 4 samples in the other two sea areas, respectively. In addition, the diversity of eukaryotic algae per sample was higher in the Laoshan Coast as well, and the top two highest diverse samples (L6 and L1) were all located in the Laoshan Coast (Fig. 7b).
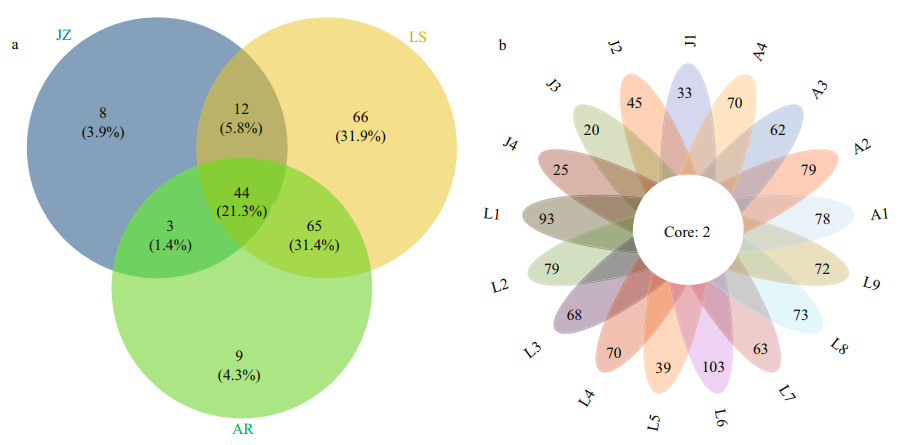
|
| Fig.7 Venn diagrams highlighting the degree of overlap of algal OTUs among the three sea areas (a) and among the seventeen samples (b) JZ: the inner Jiaozhou Bay; LS: the Laoshan Coast; AR: the Amphioxus Reserve area. |
The OTU richness, Chao1, ACE, Pielou, Shannon and Margalef indices were significantly lower in the inner Jiaozhou Bay than those in samples from the other two sea areas (P < 0.05 for all), however the Simpson index was higher in the inner Jiaozhou Bay (Supplementary Table S2). The results suggested lower diversity and evenness, and higher dominance in the benthic eukaryotic algal community in the inner Jiaozhou Bay. Stations J3 and J4, located in the eastern part of the inner Jiaozhou Bay and close to Qingdao City, had the lowest diversity, and DNA reads and proportions of algal sequences were low in the two stations as well. The small phagotrophic chrysophyte Paraphysomonas lucasi was predominated at station J1, which contributed to 96.59% of the algal sequences, leading to very low Shannon and Pielou indices and high Simpson index at the station (Supplementary Table S2). There was no significant difference in diversity indices between samples from the Laoshan Coast and the Amphioxus Reserve area (P > 0.05). The overall diversity indices in the Laoshan Coast were higher than those in the Amphioxus Reserve area due to more samples analyzed in the Laoshan Coast.
Cluster analysis showed that thirteen samples from the Laoshan Coast (L1–L9) and the Amphioxus Reserve area (A1–A4) were clustered into one large group (Fig. 8). Two samples from the inner Jiaozhou Bay (J1 and J2) were clustered into one group, while J3 and J4 were ungrouped. The results indicated the benthic algal communities were quite different between the inner Jiaozhou Bay and the other two sea areas, and also among different stations in the inner Jiaozhou Bay. Jiaozhou Bay is semi-enclosed sea area with only a narrow mouth open to the Yellow Sea, which makes it a specific ecosystem and microalgal community. While the Laoshan Coast and the Amphioxus Reserve area are connected to the Yellow Sea, which share similar microalgal compositions.
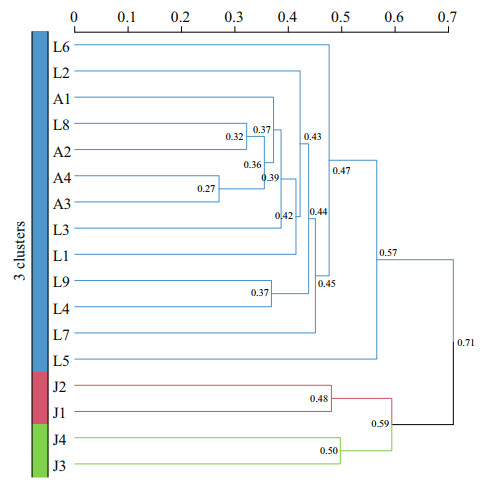
|
| Fig.8 Cluster analysis of seventeen sediment samples from the three sea areas of Qingdao coast based on Bray-Curtis distance |
Figure 9 illustrates the DNA reads of the top 25 abundant OTUs. Most of the top abundant OTUs belong to dinoflagellates, except for two OTUs in Chrysophyceae, one OTU in diatoms, and two OTUs in Raphidophyceae. Most of the identified species with high abundance were those confirmed to form resting stages (Supplementary Table S1), while those unidentified at the genus level were cyst/spore forming genera such as Gonyaulax and Chaetoceros. Only two OTUs were present at all stations (Biecheleria halophila, OTU443, OTU1061), four OTUs occurred at 16 stations, i.e., Scrippsiella acuminata (OTU1441), Gyrodinium spirale (OTU994), Gymnodinium microreticulatum (OTU1001), and Heterosigma akashiwo (OTU1455). In addition, most of the other dominant OTUs appeared at more than half of the stations. Two dinoflagellate OTUs (OTU737, OTU874) occurred abundantly in a few samples however only classified into the class lever.
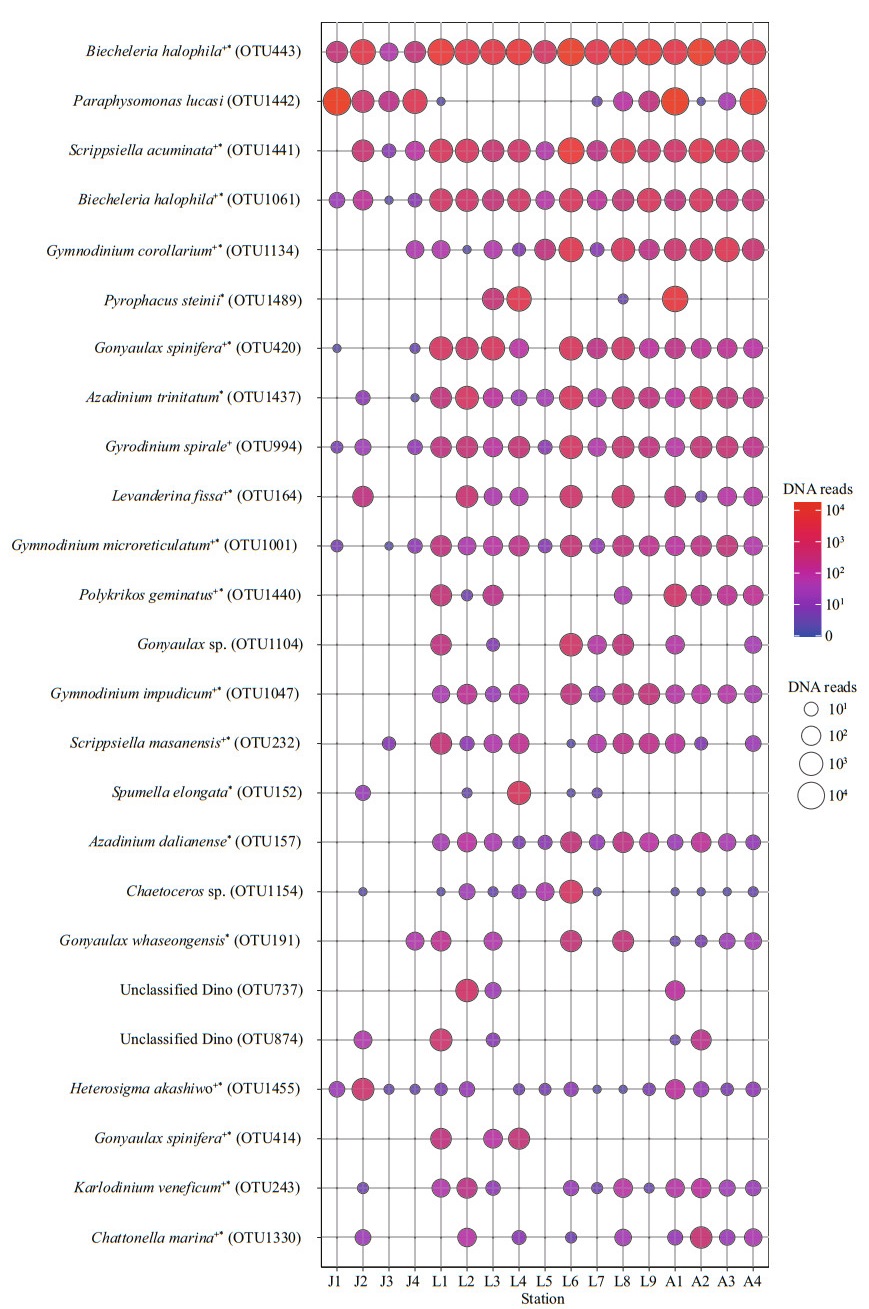
|
| Fig.9 The bubble chart showing DNA reads of the top 25 abundant OTUs in each station +: HAB species; *: taxa reported to form resting stages. Supporting evidence to HAB species and formation of resting stages are listed in Supplementary Table S1. |
The small thin-walled and cyst-forming dinoflagellate, Biecheleria halophila, was the first dominant species in the surface sediments from the Qingdao coastal areas. Two OTUs of B. halophila (OTU443 and OTU1061) were recorded in this study based on 97% similarity, which were the first and fourth dominant OTUs, respectively, and both OTUs were presented abundantly at all stations (Fig. 9). Meanwhile Biecheleria was the most diverse genus in this study, and altogether eighteen OTUs were detected into Biecheleria, ten of which were detected into four species (B. brevisulcata, B. cincta, B. halophila, and B. tirezensis). B. tirezensis was firstly recorded in the Chinese coastal waters (Supplementary Table S1).
The phagotrophic chrysophyte Paraphysomonas lucasi, a newly recorded species in Chinese coasts, was the second dominant species in this study. It was detected in 12 stations in this study and was the predominant species in the inner Jiaozhou Bay and two stations in the Amphioxus Reserve area, especially at station J1, in which accounted for 96.93% of the total algal reads. The massive occurrence of Paraphysomonas lucasi resulted in chrysophytedominant algal community in the inner Jiaozhou Bay, and chrysophyte and dinophyte co-dominant community in the Amphioxus Reserve area (Fig. 6a).
Scrippsiella acuminata (formerly S. trochoidea) was the third richest and most widely distributed species in this study. Scrippsiella is a dinoflagellate genus, which forms calcareous cysts (Matsuoka and Fukuyo, 2000). Four OTUs (OTU1441, OTU1122, OTU232, and OTU1108) were classified into genus Scrippsiella, and three of them were identified at the species level (S. acuminata, S. erinaceus, and S. masanensis, Supplementary Table S1).
Gymnodinium was one of the most common dominant genera. Five OTUs were classified into genus Gymnodinium in this study, and four were identified at the species level (Gy. catenatum, Gy. corollarium, Gy. impudicum, and Gy. microreticulatum). All these species form microreticulate cysts, which are difficult to be identified under microscopic observations (Gu et al., 2013a). Three species (Gy. corollarium, Gy. microreticulatum, and Gy. impudicum) were recorded as the top 25 abundant OTUs (Fig. 9).
Gonyaulax was one of the most diverse and abundant genera in this study. Most species of Gonyaulax form ovoid cysts with numerous membranaceous processes, which belong to palynological genus Spiniferites. Thirteen OTUs were attributed to Gonyaulax, and six OTUs were identified into four species (Go. cochlea, Go. fragilis, Go. spinifera, and Go. whaseongensis). Four OTUs including Go. spinifera (OTU420, OTU414), Gonyaulax sp. (OTU1104), and Go. whaseongensis (OTU191) were recorded as the top 25 abundance.
Azadinium occurred dominantly in this study as well, and three of the four OTUs of Azadinium were detected at the species level, i.e., Az. dalianense, Az. poporum, and Az. trinitatum. Az. trinitatum (OTU1437) and Az. dalianense (OTU157) were the top 8th and top 17th abundant OTUs, respectively. In addition, Az. trinitatum distributed in 15 stations, and Az. dalianense was recorded in all stations except for four stations in the inner Jiaozhou Bay. While the azaspiracid shellfish poisoning (AZP) producer Az. poporum (Krock et al., 2012) occurred in only three samples with low reads.
Chaetoceros was the most abundant and diverse diatom genus in this study, and eight OTUs were detected including six OTUs identified into five species (Chaetoceros debilis, Chae. decipiens, Chae. muelleri, Chae. radicans, and Chae. wighamii). Only one diatom OTU (Chaetoceros sp., OTU1154) was within the top 25 abundances, and other diatoms occurred in low reads.
3.5 Occurrence of harmful algal bloom speciesNotably, 36 potentially toxic/harmful and/or bloom species were detected in surface sediments from the Qingdao coast (Supplementary Table S1), including the paralytic shellfish poisoning (PSP) producers Alexandrium minutum, Al. andersonii, and Gymnodinium catenatum, the yessotoxin (YTX) producers Gonyaulax spinifera and Protoceratium reticulatum, the AZA producer Azadinium poporum, the karlotoxin producer Karlodinium veneficum, the ichthyotoxic species Aureococcus anophagefferens, Chattonella marina, Fibrocapsa japonica, Heterosigma akashiwo, Polykrikos hartmannii, and Po. geminatus. Sixteen bloom species occurred in all of the three sea areas, including three bloom causative diatoms, ten HAB dinoflagellates, the brown tide species Au. anophagefferens, and two ichthyotoxic raphidophytes Chat. marina and H. akashiwo. All of the 36 bloom species were recorded in the Laoshan Coast, and 20 and 27 bloom species were detected in samples from the inner Jiaozhou Bay and the Amphioxus Reserve area, respectively. Some of these species occurred widely and dominantly in this study, such as B. halophila, S. acuminata, Gy. corollarium, Go.spinifera, Levanderina fissa, Gy.microreticulatum, Po. geminatus, Gy. impudicum, H. akashiwo, K. veneficum, and Chat. marina (Fig. 9).
3.6 RDA analyses of benthic algal community and biogenic elementsRDA analysis was conducted based on biogenic elements and benthic eukaryotic algal community (Fig. 10). Axis 1 and Axis 2 explained 61.9% and 20.7% of the environmental and biological variables, respectively. Most of the biogenic elements and eukaryotic algal classes were located in the negative direction of Axis 1, and BSi had a higher negative contribution to Axis 1. Dinoflagellates (Dinophyceae and Syndiniophyceae) and diatoms (Bacillariophyceae and Mediophyceae) showed narrow intersection angles with BSi and TP, indicating significant influences of the two biogenic elements on the distribution of dinoflagellates and diatoms (Fig. 10a). TOC, OM, and TN had a negative contribution to Axis 2. Two classes of green algae, Pyramimonadophyceae and Mamiellophyceae, had a small angle with these elements, which indicated that organic matters had a certain influence on the distribution of green algae. Other algal classes scattered in the coordinates far away from the biogenic elements, indicating few effects of biogenic elements on their distribution.

|
| Fig.10 Redundancy analysis (RDA) of biogenic elements and the eukaryotic algal community in surface sediments from the Qingdao coasts, showing algal distribution on class level and the directions of biogenic elements to the first two RDA axes, axis 1 (61.9%) and axis 2 (20.7%) The length of arrows indicates the importance of biogenic elements in explaining the distribution of eukaryotic algae. The direction of arrow shows approximate correlation to the ordination axes. a. eukaryotic algae in class level; b. ordination sampling stations, showing the distribution of the three areas; TOC: total organic carbon; OM: organic matter; TN: total nitrogen; TP: total phosphorus; BSi: biogenic silicon. Classes are represented in (a) by their first four characters the same as shown in Fig. 4. |
The ordination sampling station showed that samples in the three sea areas were separately grouped (Fig. 10b). Samples in the inner Jiaozhou Bay were clustered along the positive axis of Axis 1, which indicated the low BSi content in this area. Samples in the Amphioxus Reserve area distributed along the positive axis of Axis 2, indicating the low organic matters (TOC, OM, and TP) in this area. Meanwhile, the arrows of the biogenic elements distributed within the group of samples of the Laoshan Coast, indicating the high contents of biogenic elements in this area. Station J3 was far away from other stations, which might be accounted by the influences of intensive human activities as it is very close to land. Furthermore, only 424 algal reads were recorded in J3, which was too low to form a strong correlation with biogenic elements.
4 DISCUSSION 4.1 Composition of eukaryotic algal community in surface sediments from the Qingdao coastDinoflagellates dominated in the benthic algal community structure in this study, followed by chrysophytes, diatoms, and chlorophytes. Dinoflagellates also had the highest OTU richness. The benthic algal community structure is comparable to those reported in sediments from the other coastal sea areas, which were dominated by dinoflagellates in both DNA reads and OTU richness (Dzhembekova et al., 2018; Liu et al., 2020a). Dinoflagellates have much higher DNA copies (up to thousands) than the other protists (dozens to hundreds) (Lin, 2011), which may lead to their over-representation in metabarcoding studies. In addition, about 20% dinoflagellates have the ability to form resting cysts, and dinoflagellate cysts constitute a major component of sedimentary assemblages of microalgal resting stages (reviewed by Ellegaard and Ribeiro, 2018). Actually, most dominant dinoflagellates detected at the species level in this study were cyst-forming taxa (Fig. 9).
Chrysophyceae has been detected abundantly in the eukaryotic sequences in both sediment and water samples (de Vargas et al., 2015; Liu et al., 2020a). Cyst formation is a characteristic feature of chrysophytes, which form stomatocysts with siliceous wall (Findenig et al., 2010). Therefore, the chrysophyte community has been recorded relatively completely in metagenomic monitoring of sediments. Paraphysomonas lucasi predominated in the inner Jiaozhou Bay and also widely and abundantly distributed in the sediments from the Amphioxus Reserve area in this study. It is a new recorded species in the Chinese coastal sea area as far as our knowledge. Wide and massive occurrences of this species in this study suggested its ubiquitous distribution and cyst-formation characteristics. Meanwhile, chrysophytes are also an important group in phytoplankton community (Liu et al., 2020b; Pan et al., 2020). However, they are generally difficult to be distinguished and identified under the microscopic observations due to their fragility or their small size (LeRoi and Hallegraeff, 2006; Findenig et al., 2010). More and more chrysophyte species have been recorded in phytoplankton communities as the application of HTS-based metabarcoding (Scoble and Cavalier-Smith, 2014), for example Paraphysomonas foraminifera, Pa. imperforate, and Pedospumella encystans in Jiaozhou Bay, the Yellow Sea (Liu et al., 2020b), and Paraphysomonas vestita in Liaodong Bay, the Bohai Sea (Song et al., 2019). The results of these studies and our study suggested that the abundance and diversity of the nano-sized chrysophytes might be much greater than expected in the Yellow Sea and the Bohai Sea of China.
The OTU richness and abundance of benthic diatoms were greatly reduced in this study compared with the phytoplankton metabarcoding studies in Jiaozhou Bay and the other sea areas (Xu et al., 2017; Song et al., 2019; Liu et al., 2020b). We focused on phytoplankton resting stages in this study. However, only a few central diatom species have been reported to form resting spores (McQuoid and Hobson 1996). Therefore, most diatom cells existing in water column could not be detected in this study. Diatoms recorded in this study were those reported to form resting spores, such as species in genera Chaetoceros, Skeletonema, and Thalassiosira etc. (Montresor et al., 2013; Piredda et al., 2017). Meanwhile, the abundance and species diversity of diatoms in the phytoplankton community in Jiaozhou Bay and its adjacent waters showed a decreasing trend due to the decrease of silicon flux (Liu et al., 2010). Some freshwater and terrestrial species were detected in this study, which might be carried into the sea by surface runoff.
4.2 Comparison of benthic algal communities in the three sea areasAlthough the three sea areas are not far apart, the benthic algal community structures were quite different. Chrysophytes and dinoflagellates dominated in the inner Jiaozhou Bay and the Laoshan sea area, respectively. While, dinoflagellates and chrysophytes co-dominated in the Amphioxus Reserve area. Only 44 OTUs (21.3%) shared among the three sea areas, and 2 OTUs shared among all samples in this study. The cluster analysis showed that samples from Jiaozhou Bay were separately clustered. Jiaozhou Bay is an enclosed inner bay with little exchange with outside water bodies. The four stations of Jiaozhou Bay in this study are located in the inner Jiaozhou Bay. The enclosed environment results in a unique phytoplankton and benthic algal community structure. The predominant chrysophyte Paraphysomonas lucasi recorded in this study is a ubiquitous phagotroph, which primarily feeds on bacteria (Scoble and Cavalier-Smith, 2014). Therefore, nutrient enrichment and high organic matters in Jiaozhou Bay (Li et al., 2020) provide suitable conditions for the phagotrophic chrysophytes. Actually, chrysophytes occurred abundantly in the phytoplankton community in Jiaozhou Bay as well, and the proportion of chrysophytes was greater in the inner Jiaozhou Bay (Liu et al., 2020b).
Two hundred and seven eukaryotic algae OTUs were obtained from 17 sediment samples from the Qingdao coast in this study, which was comparable to other metabarcoding studies on dinoflagellate cysts and phytoplankton resting stages (Dzhembekova et al., 2018; Liu et al., 2020a). However, compared with benthic microeukaryote studies in the Yellow Sea and European coastal areas (Gong et al., 2015; Massana et al., 2015), lower diversity of eukaryotic algae community was recorded in Qingdao coastal waters in this study. This study focused on the phytoplankton resting stages, and samples were stored at 4 ℃ for 6 months to inactivate most of the vegetative cells (Piredda et al., 2017). Only some parts of dinoflagellates, diatoms, chrysophytes, chlorophytes, haptophytes, and raphidophytes form resting stages, which can stay alive in sediments for a long time (reviewed by Ellegaard and Ribeiro, 2018). In addition, the inner Jiaozhou Bay had the lowest diversity and algal sequence number among the three sea areas, especially at station J3, in which only ca. 400 algal reads were recorded. While eukaryotic sequence numbers measured in all the four samples from Jiaozhou Bay were more than 50 000 reads (Fig. 2) with sufficient sequencing depth. Eutrophication in Jiaozhou Bay has greatly increased recently, and the nutrient status has changed, which resulted in frequent algal blooms and decreased phytoplankton biodiversity (Shi et al., 2020). In addition, Jiaozhou Bay is an important sea area for shellfish culturing, and the feeding of shellfish reduces the diversity of eukaryotic microalgae to a certain extent (Yu et al., 2019).
4.3 Neglected taxa under routine microscopic observationOne remarkable difference between our study and the previous traditional phytoplankton and resting stage studies is that diverse phyla and classes were recorded in this study, including 24 classes of 8 phyla. However, diatoms and dinoflagellates were mostly recorded in the previous phytoplankton surveys based on traditional microscopic observations, and other phyla such as Chlorophyta, Cryptophyta, Haptophyta, Ochrophyta, Prasinodermatophyta, and Rhodophyta were rarely reported (Yang et al., 2014; Guo et al., 2019). Metabarcoding has a strong ability to detect the small-sized, morphologically similar, and the fragile and rare species (Salonen et al., 2019). A large number of small-sized species were detected in this study, for example, species in genera Azadinium, Biecheleria, and Paragymnodinium (< 20 μm), calcareous nannofossils Achradina pulchra (< 20 μm) and Braarudosphaera bigelowii (< 5 μm), plenty of nano-sized chlorophytes and chrysophytes (< 10 μm) (Supplementary Table S1). Liu et al. (2020b) analyzed phytoplankton community in Jiaozhou Bay based on metabarcoding, and detected 70 species which had never been reported in Jiaozhou Bay in the previous investigations, and majority of these species missed by previous studies are relatively small in size (< 10 μm), mostly belonging to Cryptophyta, Chlorophyta, Ochrophyta, and Haptophyta, which are difficult to detect and identify using light microscopy. Meanwhile, Pigments from eight possible phytoplankton classes (Diatoms, Dinoflagellates, Chlorophyceae, Prasinophyceae, Chrysophyceae, Haptophyceae, Cryptophyceae, and Cyanobacteria) were detected in surface water by high performance liquid chromatography (HPLC) (Yao et al., 2010). These studies also suggested high diversities of phytoplankton community in Jiaozhou Bay.
Twelve taxa are firstly recorded in the Chinese coastal waters in this study after reviewing the literatures based on both microscopic and metabarcoding studies (Supplementary Table S1). Many new recorded species occurred abundantly and widely in this study, and might be overlooked in previous microscopic studies due to the small size, fragile cell wall, and indistinguishable morphological characteristics. Furthermore, remarkable diverse chlorophytes were recorded in our study, including 34 OTUs in 10 classes. Most of these chlorophytes were recorded in previous phytoplankton metabarcoding studies (Xu et al., 2017; Song et al., 2019; Liu et al., 2020b). Three species in chlorophytes together with the other nine species have not been detected in previous phytoplankton metabarcoding studies in Chinese coasts (Xu et al., 2017; Song et al., 2019; Liu et al., 2020b), however were recorded in our study on sediments. It is well known that the resting stages in sediments are the assemblages of the deposition of phytoplankton resting stages over a period, while results of phytoplankton surveys reflect the phytoplankton community in the water column at the sampling moment. Actually, many these new recorded species are resting stage forming or have the thick calcareous wall, which can be the fossil assemblages in sediments (Supplementary Table S1).
4.4 Occurrence of HAB species and potential risks of HABs in the Qingdao coastThirty-six potentially toxic/harmful and/or bloom species were detected in surface sediments from the Qingdao coast, among which ten species, including Gonyaulax spinifera, Noctiluca scintillans, Prorocentrum micans, Scrippsiella acuminata, Aureococcus anophagefferens, Heterosigma akashiwo, Chattonella marina (Ministry of Ecology and Environment of the People's Republic of China, 2010–2017), Chaetoceros debilis (Yan et al., 2002), Margalefidinium fulvescens (Lin et al., 2020), and Nannochloropsis granulate (Zhang et al., 2015), have been reported to form blooms in Jiaozhou Bay and the Qingdao coast before. Some of them are toxic species, which may produce toxins penitently harmful to human health. Most HAB species found in this study can form resting stages (29 to 36 species, Supplementary Table S1), and some of them occurred widely and abundantly in the sediments (Fig. 9). Notably all of the 36 HAB species occurred in sediment samples from the Laoshan Coast. The result suggested the high potential risk of HABs in the Qingdao coastal area, especially in the Laoshan Coast.
Dinoflagellates were the most important HAB group identified in this study. Twenty-two species of dinoflagellates were identified to produce toxins and/or form blooms (Supplementary Table S1). The potential PSP producers, Alexandrium andersonii, Al. minutum, and Gymnodinium catenatum (Lundholm et al., 2009 onwards), were present in sediments but in low DNA reads, indicating that the low historical abundance of these species in the water column. Liu et al. (2020b) did not identify any of these PSP producers in the phytoplankton survey in January 2019. The potential YTX producer, Gonyaulax spinifera and Protoceratium reticulatum (Lundholm et al., 2009 onwards), were detected in this study. Three OTUs were attributed to G. spinifera, two of which belonged to the top abundant OTUs (Fig. 9). Blooms of G. spinifera occurred at outside of Jiaozhou Bay in 2003 and 2007, respectively (Zhou et al., 2020). As a cyst-forming species, G. spinifera can form variety types of cysts, mostly belonging to the cyst genus Spiniferites with wide morphological variations (Rochon et al., 2009). The great occurrence of G. spinifera in the Laoshan Coast (Fig. 9) indicated high numbers of its vegetative cells in the water column and the high risk of its blooms. Another Ganyaulax species, G. fragilis which has been associated with the massive mucilage formations in the Marmara Sea, Turkey (Balkis et al., 2011), rarely distributed at one station of the Laoshan Coast in this study.
Three species of Azadinium were detected in this study, only Az. poporum has the potential to produce AZP (Krock et al., 2012). Az. poporum was reported to distribute widely in surface sediments from the Chinese coasts (Gu et al., 2013b, Liu et al., 2020a), and 13 out of 16 strains of Az. poporum from different geographic locations along the Chinese coastline contained azaspiracids (Krock et al., 2014). DNA sequence number of Az. poporum was relatively low, on the other hand, the nontoxic Az. trinitatum and Az. dalianense widely distributed in high DNA reads in this study (Fig. 9).
The karlotoxin producer, Karlodinium veneficum, occurred in most samples (Fig. 9). K. veneficum is a small sized naked doniflagellate, which produces a groups of polyketide toxins (karlotoxins) exerting hemolytic, ichthyotoxic, and cytotoxic effects on fishes and aquatic animals (Place et al., 2012). Since the first bloom of K. veneficum reported in the South China Sea in 2003 (Liu et al., 2020c), its blooms occurred in the Chinese coastal sea areas frequently (Wang et al., 2011; Dai et al., 2014). Cysts of this species were detected widely in sediments from the China Sea as well (Liu et al., 2020c). Considering the association of its blooms and massive fish killings (Place et al., 2012), the wide distribution of K. veneficum is worthy of attention.
Biecheleria was rarely observed in the previous routine phytoplankton surveys due to the small size and frangible thin wall. Biecheleria has become more frequently detected in the phytoplankton communities as the development of molecular biological techniques (Sundström et al., 2010). Moreover, taxa in Biecheleria generally form resting cysts (Moestrup et al., 2009), which made them as the common dominant eukaryotes in sediments (Dzhembekova et al., 2018; Liu et al., 2020a; Rhodes et al., 2020). Ten OTUs of Biecheleria were detected in sediments from the Qingdao coast, and B. halophila was the first dominant species detected in this study and B. tirezensis was newly recorded in the Chinese coastline. B. halophila has been reported to form a co-occurring bloom with Scrippsiella hangoei in the Baltic Sea (Kremp et al., 2005). The results suggested that species in Biecheleria were probably of much wider distribution, however might be ignored during the microscopic observation because of their fragility and small size.
Other ichthyotoxic species, whose blooms have been associated with massive fish kills, included the dinoflagellates Polykrikos hartmannii, and Po. geminatus, the pelagophyte Aureococcus anophagefferens, and the raphidophytes Chattonella marina, Fibrocapsa japonica, and Heterosigma akashiwo (Supplementary Table S1). Among them, blooms of Au. anophagefferens, Chat. marina, and H. akashiwo occurred in Jiaozhou Bay and the nearby sea areas (Ministry of Ecology and Environment of the People's Republic of China, 2010–2017), and brown tides caused by Au. anophagefferens have recurred frequently in the northwestern Bohai Sea since 2009 (Huang et al., 2020). Po. geminatus is a common bloom species in the Pearl River Estuary of the South China Sea, and its blooms have frequently occurred since 2006 (Shen et al., 2012; Dong et al., 2020). Po. geminatus was detected in sediment samples from eight stations in this study, and ranked the top 12th abundant OTU (Fig. 9). To the best of our knowledge, it is the first report of this species present in the northern Chinese coastline, i.e., the Yellow sea and the Bohai Sea. Coincidentally, another bloom species in the Pearl River Estuary, Levanderina fissa, occurred widely and abundantly in this study (Fig. 9).
4.5 Potential of metabarcoding for microalgal resting stage monitoringMicroscopic observations require professional taxonomic expertise for distinguishing the tiny morphological differences among species from an assemblage of organisms (Shang et al., 2019). Furthermore, only a small proportion of phytoplankton resting stages are known for their corresponding vegetative cells, and vice versa. Misidentification is therefore common and almost unavoidable due to these difficulties (Gómez et al., 2017; Shang et al., 2019). Metabarcoding can avoid these problems, and standardized protocols can be developed to make results more comparable among laboratories (Pochon et al., 2015). Metabarcoding has the ability to detect small-sized taxa that were not observed in previous microscopic surveys, such as species in Azadinium, Biecheleria, and Paragymnodinium, calcareous nannofossils Achradinapulchra and Braarudosphaera bigelowii, and the nano-sized chlorophytes in this study. The results suggest that metabarcoding provides comprehensive insights into benthic communities, especially for organisms that are difficult to identify because of their small size, cryptic habits (such as the parasitic and symbiotic species), occurrence in the form of resting stages or lack of traditional identification keys (Salonen et al., 2019).
The first problem in sediment metabarcoding analysis is the less efficiency of genomic DNA extraction for resting stages than for vegetative cells, due to the presence of the thick resistant walls. In addition, DNA extraction is particularly difficult from sediment samples containing high concentrations of clay and humic material. Therefore, extraction and purification of DNA are crucial steps in the analysis of marine sediments, where the presence of high organic matter, organic pollutants and heavy metals can interfere with the recovery of DNA and reduce the effectiveness of PCR (Piredda et al., 2017). This might be part of the reasons the lower diversity in the sediment sample than in phytoplankton. We applied the standardized protocol suggested by DNeasy PowerSoil Kit for DNA isolation, which included complete vortex of the soil samples in the PowerBead tube with Solution C1 to break the tough cells such as resting stages. The kit has been widely used and shown to be well optimized for DNA extraction from a variety of soils and plant tissues (reviewed by Lear et al., 2018). Actually, high diversity of eukaryotic algae (207 OTUs) was obtained in this study, and the cyst/spore-forming taxa such as the dinoflagellates Biecheleria, Scrippsiella, Gymnodinium, the Chrysophyte Paraphysomonas, and the diatom Chaetoceros were the dominant ribotypes in the sediments. The results indicated that DNA was effectively extracted from these thick-wall spores/cysts, and thus suggested the efficiency of DNA extraction.
Our samples were stored in a cold dark condition for six months in order to ensure that most survives in the sediments were resting stages (Piredda et al., 2017). About half taxa that were identified to the species level in this study have been reported to form resting stages (Supplementary Table S1), and most of the top abundant OTUs were reported to form resting stages (Fig. 9). Other taxa such as Paraphysomonas lucasi, Gonyaulax sp., and Chaetoceros sp. occurred abundantly in this study (Fig. 9). However, whether they could form resting stages was still unknown. Indeed, cyst/spore formation is an important characteristic in genus Paraphysomonas (Coradeghini and Vigna, 2008), Gonyaulax (Matsuoka and Fukuyo, 2000), and Chaetoceros (Montresor et al., 2013). Microscopic observations were undertaken to observe dinoflagellate cysts in the same sediments synchronously, and 23 taxa of dinoflagellate cysts were observed, and only 13 taxa were identified to the species level (Lei, 2021). While 127 dinoflagellate OTUs were detected by metabarcroding in this study, and 66 OTUs were attributed to 45 species of dinoflagellates, including 32 cyst-forming species (Supplementary Table S1). The results suggested much higher biodiversity of dinoflagellate cysts was revealed by metabarcroding than that by microscopy. However, we did not analyze resting stages of the other phytoplankton groups by microscopy due to the lack of morphological references. Those taxa that do not produce resting stages were generally detected in low DNA reads, some of which might be the benthic vegetative cells, and some of which might represent DNA fragments of dead cells or even DNA relics. Actually, it is hard to distinguish the dead and partially degraded phytoplankton cells from authentic sediment inhabiting cells or resting cells during HTS based metabarcoding, which may increase the OTU richness to some extent. However, DNA fragments would be presented as extremely rare OTUs after long time storage under cold dark condition (Shang et al., 2019). The rare OTUs with 1–2 reads (singletons and doubletons) were removed in this study, which reduced the possibility of increased OTU richness due to DNA fragments. On the other hand, storing sediment samples for six months could turn benthic microalgae into resting stages, which might increase OTU richness and diversity of microalgal resting stages in our study. However, majority of cyst/spore forming taxa are phytoplankton species (reviewed by Ellegaard and Ribeiro, 2018), and only a few benthic algae have been reported to form resting stages up until now (Limoges et al., 2015; Mertens et al., 2017). Furthermore, most of the species identified in this study were marine phytoplankton (Supplementary Table S1). Therefore, the benthic algae had little contribution to the diversity in this study.
The choice of the targeted region has a potentially significant influence in metabarcoding surveys. The V4 and V9 regions of 18S rDNA are two of the most commonly used regions for metabarcoding eukaryotic organisms. The V4 region is the longest 18S region in eukaryotes and features high variability, which increases its resolution relative to other 18S rRNA regions (reviewed by Lear et al., 2018). The V4 region is one of the best choices for metabarcoding the entire eukaryotic community from environmental DNA (reviewed by Lear et al., 2018) and has been used in many studies focusing on marine microalgae (Kataoka et al., 2017; Penna et al., 2017; Xu et al., 2017; Song et al., 2019; Liu et al., 2020b; Cui et al., 2021). Gaonkar et al. (2018) compared the 18S rDNA V4 and V9 hypervariable region sequences of taxa in the planktonic diatom family Chaetocerotaceae, and found that the 18S V4 region fit well with target taxa, and the V4-tree topology was similar to that of the whole 18S rDNA. The primer sites used in our study produce amplicons of lengths (450 bp) just right ideal length for sequencing on the Illumina MiSeq platform. Results from the phylogenetic tree showed that OTUs in the same phylum were generally clustered together (Fig. 5), suggesting the credibility of species annotation.
In addition, the reference database used potentially has a large impact on the eukaryote community obtained. Of the 207 generated algal OTUs, only 124 OTUs (59.9%) were identified at the species level, and some unclassified OTUs were the common predominant sequences in our study. The lack of coverage at the species level for many taxa may hinder correct interpretation of metabarcoding data (Forster et al., 2016). However, as new sequencing data are continuously contributing to the existing databases, the situation is likely to improve in the future. On the other hand, the identification and enumeration of microalgae based on microscopic methods are restricted to organisms with well-documented morphological characteristics (Piredda et al., 2017). While the morphologies of resting stages of most microalgae are still unknown, and it is difficult to analyze them under microscopy. The capacity of HTS to detect morphologically cryptic, small-sized, and rare taxa makes this method a sufficiently sensitive detection approach for assessing the composition of resting stages in sediments.
5 CONCLUSIONThis study enriched our understanding of eukaryotic algae in surface sediments from the Qingdao Coast based on 18S rDNA metabarcoding. Diverse eukaryotic algae were recorded, including 207 OTUs belonged to 75 genera in 24 classes of 8 phyla. Within 98 taxa identified to the species level, 12 species were firstly recorded in the Chinese coastal waters. Dinophyta generally dominated in both DNA reads and OTU richness. The benthic algal community structures were quite different in three sea areas of the Qingdao coast, which was dominated by chrysophytes in Jiaozhou Bay, by dinoflagellates in the Laoshan Coast, and co-dominated by dinoflagellates and chrysophytes in the Amphioxus Reserve area. Furthermore, eukaryotic algae OTU richness and diversity were higher in the Laoshan Coast, and lower in Jiaozhou Bay. A large number of small-sized species were detected in this study, such as Azadinium spp., Biecheleria spp., and Paragymnodinium spp., and nano-sized chlorophytes and chrysophytes. The results suggested the strong ability of metabarcoding to detect the small-sized, morphologically similar, and the fragile and rare species. Notably, 36 HAB species were detected in this study, and some of these species occurred widely and dominantly in this study, suggesting high potential risk of HABs in the Qingdao coastal area.
6 DATA AVAILABILITY STATEMENTThe data in this study are available from the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/Traces/sra) with the accession number PRJNA722324.
Electronic supplementary material
Supplementary material (Supplementary Tables S1–S2) is available in the online version of this article at https://doi.org/10.1007/s00343-021-1200-0.
Balkis N, Atabay H, Türetgen I, Albayrak S, Balkis H, Tüfekçi V. 2011. Role of single-celled organisms in mucilage formation on the shores of Büyükada Island (the Marmara Sea). Journal of the Marine Biological Association of the United Kingdom, 91(4): 771-781.
DOI:10.1017/S0025315410000081 |
Bik H M, Sung W, De Ley P, Baldwin J G, Sharma J, Rocha-Olivares A, Thomas W K. 2012. Metagenetic community analysis of microbial eukaryotes illuminates biogeographic patterns in deep-sea and shallow water sediments. Molecular Ecology, 21(5): 1048-1059.
DOI:10.1111/j.1365-294X.2011.05297.x |
Blanco A C, Nadaoka K, Yamamoto T. 2008. Planktonic and benthic microalgal community composition as indicators of terrestrial influence on a fringing reef in Ishigaki Island, Southwest Japan. Marine Environmental Research, 66(5): 520-535.
DOI:10.1016/j.marenvres.2008.08.005 |
Bråte J, Logares R, Berney C, Ree D K, Klaveness D, Jakobsen K S, Shalchian-Tabrizi K. 2010. Freshwater Perkinsea and marine-freshwater colonizations revealed by pyrosequencing and phylogeny of environmental rDNA. The ISME Journal, 4(9): 1144-1153.
DOI:10.1038/ismej.2010.39 |
Caporaso J G, Kuczynski J, Stombaugh J, Bittinger K, Bushman F D, Costello E K, Fierer N, Peña A G, Goodrich J K, Gordon J I, Huttley G A, Kelley S T, Knights D, Koenig J E, Ley R E, Lozupone C A, Mcdonald D, Muegge B D, Pirrung M, Reeder J, Sevinsky J R, Turnbaugh P J, Walters W A, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nature Methods, 7(5): 335-336.
DOI:10.1038/nmeth.f.303 |
Cavalier-Smith T, Lewis R, Chao E E, Oates B, Bass D. 2009. Helkesimastix marina n. sp. (Cercozoa: Sainouroidea superfam. n. ) a gliding zooflagellate of novel ultrastructure and unusual ciliary behaviour.. Protist, 160(3): 452-479.
DOI:10.1016/j.protis.2009.03.003 |
Chen T T, Zhang Y X, Song S Q, Liu Y, Sun X X, Li C W. 2021. Diversity and seasonal variation of marine phytoplankton in Jiaozhou Bay, China revealed by morphological observation and metabarcoding. Journal of Oceanology and Limnology, published online on July 2021, https://doi.org/10.1007/s00343-021-0457-7.
|
Coradeghini A, Vigna M S. 2008. The aerial epiphytic stomatocyst flora (Chrysophyceae and Synurophyceae) of mosses from Primavera Station, Antarctica. Nova Hedwigia, 86(3-4): 401-417.
DOI:10.1127/0029-5035/2008/0086-0401 |
Cui Z M, Xu Q, Gibson K, Liu S Y, Chen N S. 2021. Metabarcoding analysis of harmful algal bloom species in the Changjiang Estuary, China. Science of the Total Environment, 782: 146823.
DOI:10.1016/j.scitotenv.2021.146823 |
Dai X F, Lu D D, Guan W B, Wang H X, He P X, Xia P, Yang H J. 2014. Newly recorded Karlodinium veneficum dinoflagellate blooms in stratified water of the East China Sea. Deep Sea Research Part II: Topical Studies in Oceanography, 101: 237-243.
DOI:10.1016/j.dsr2.2013.01.015 |
de Vargas C, Audic S, Henry N, Decelle J, Mahé F, Logares R, Lara E, Berney C, Bescot N L, Probert I, Carmichael M, Poulain J, Romac S, Colin S, Aury J M, Bittner L, Chaffron S, Dunthorn M, Engelen S, Flegontova O, Guidi L, Horák A, Jaillon O, Lima-Mendez G, Lukeš J, Malviya S, Morard R, Mulot M, Scalco E, Siano R, Vincent F, Zingone A, Dimier C, Picheral M, Searson S, KandelsLewis S, Coordinators T O, Acinas S G, Bork P, Bowler C, Gorsky G, Grimsley N, Hingamp P, Iudicone D, Not F, Ogata H, Pesant S, Raes J, Sieracki M E, Speich S, Stemmann L, Sunagawa S, Weissenbach J, Wincker P, Karsenti E, Boss E, Follows M, Karp-Boss L, Krzic U, Reynaud E G, Sardet C, Sullivan M B, Velayoudon D. 2015. Eukaryotic plankton diversity in the sunlit ocean. Science, 348(6237): 1261605.
DOI:10.1126/science.1261605 |
Dong Y L, Cui L, Cao R B, Cen J Y, Zou J, Zhou X Y, Lu S H. 2020. Ecological characteristics and teratogenic retinal determination of Cochlodinium geminatum blooms in Pearl River Estuary, South China. Ecotoxicology and Environmental Safety, 191: 110226.
DOI:10.1016/j.ecoenv.2020.110226 |
Dzhembekova N, Moncheva S, Ivanova P, Slabakova N, Nagai S. 2018. Biodiversity of phytoplankton cyst assemblages in surface sediments of the Black Sea based on metabarcoding. Biotechnology & Biotechnological Equipment, 32(6): 1507-1513.
DOI:10.1080/13102818.2018.1532816 |
Ellegaard M, Ribeiro S. 2018. The long-term persistence of phytoplankton resting stages in aquatic 'seed banks'. Biological Reviews, 93(1): 166-183.
DOI:10.1111/brv.12338 |
Fierro P, Valdovinos C, Arismendi I, Díaz G, Jara-Flores A, Habit E, Vargas-Chacoff L. 2019. Examining the influence of human stressors on benthic algae, macroinvertebrate, and fish assemblages in Mediterranean streams of Chile. Science of the Total Environment, 686: 26-37.
DOI:10.1016/j.scitotenv.2019.05.277 |
Findenig B M, Chatzinotas A, Boenigk J. 2010. Taxonomic and ecological characterization of stomatocysts of Spumella-like flagellates (Chrysophyceae). Journal of Phycology, 46(5): 868-881.
DOI:10.1111/j.1529-8817.2010.00892.x |
Forster D, Dunthorn M, Mahé F, Dolan J R, Audic S, Bass D, Bittner L, Boutte C, Christen R, Claverie J M, Decelle J, Edvardsen B, Egge E, Eikrem W, Gobet A, Kooistra W H C F, Logares R, Massana R, Montresor M, Not F, Ogata H, Pawlowski J, Pernice M C, Romac S, Shalchian-Tabrizi K, Simon N, Richards T A, Santini S, Sarno D, Siano R, Vaulot D, Wincker P, Zingone A, De Vargas C, Stoeck T. 2016. Benthic protists: the under-charted majority. FEMS Microbiology Ecology, 92(8): fiw120.
DOI:10.1093/femsec/fiw120 |
Gaonkar C C, Piredda R, Minucci C, Mann D G, Montresor M, Sarno D, Kooistra W H C F. 2018. Annotated 18S and 28S rDNA reference sequences of taxa in the planktonic diatom family Chaetocerotaceae. PLoS One, 13(12): e0208929.
DOI:10.1371/journal.pone.0208929 |
Genovesi B, Laabir M, Masseret E, Collos Y, Vaquer A, Grzebyk D. 2009. Dormancy and germination features in resting cysts of Alexandrium tamarense species complex (Dinophyceae) can facilitate bloom formation in a shallow lagoon (Thau, southern France). Journal of Plankton Research, 31(10): 1209-1224.
DOI:10.1093/plankt/fbp066 |
Gómez F, Richlen M L, Anderson D M. 2017. Molecular characterization and morphology of Cochlodinium strangulatum, the type species of Cochlodinium, and Margalefidinium gen. nov. for C. polykrikoides and allied species (Gymnodiniales, Dinophyceae). Harmful Algae, 63: 32-44.
DOI:10.1016/j.hal.2017.01.008 |
Gong J, Shi F, Ma B, Dong J, Pachiadaki M, Zhang X L, Edgcomb V P. 2015. Depth shapes α- and β-diversities of microbial eukaryotes in surficial sediments of coastal ecosystems. Environmental Microbiology, 17(10): 3722-3737.
DOI:10.1111/1462-2920.12763 |
Gu H F, Liu T T, Vale P, Luo Z H. 2013a. Morphology, phylogeny and toxin profiles of Gymnodinium inusitatum sp. nov., Gymnodinium catenatum and Gymnodinium microreticulatum (Dinophyceae) from the Yellow Sea, China. Harmful Algae, 28: 97-107.
DOI:10.1016/j.hal.2013.06.001 |
Gu H F, Luo Z H, Krock B, Witt M, Tillmann U. 2013b. Morphology, phylogeny and azaspiracid profile of Azadinium poporum (Dinophyceae) from the China Sea. Harmful Algae, 21-22: 64-75.
DOI:10.1016/j.hal.2012.11.009 |
Gu X, Xu L, Wang Z X, Ming X, Dang P, Ouyang W, Lin C Y, Liu X T, He M C, Wang B D. 2021. Assessment of cadmium pollution and subsequent ecological and health risks in Jiaozhou Bay of the Yellow Sea. Science of the Total Environment, 774: 145016.
DOI:10.1016/j.scitotenv.2021.145016 |
Guiry M D, Guiry G M. 2021. AlgaeBase. World-wide electronic publication. National University of Ireland, Galway. http://www.algaebase.org. Accessed on 2021-08-09.
|
Guo S J, Zhu M L, Zhao Z X, Liang J H, Zhao Y F, Du J, Sun X X. 2019. Spatial-temporal variation of phytoplankton community structure in Jiaozhou Bay, China. Journal of Oceanology and Limnology, 37(5): 1611-1624.
DOI:10.1007/s00343-019-8249-z |
Hallegraeff G M, Anderson D M, Cembella A D. 2003. Manual on Harmful Marine Microalgae. UNESCO Publishing, Paris. 793p.
|
Head M J. 1996. Modern dinoflagellate cysts and their biological affinities. In: Jansonius J, McGregory D C eds. Palynology: Principles and Applications. American Association of Stratigraphic Palynologists Foundation, Dallas. p. 1197-1248.
|
Huang B Q, Liang Y L, Pan H Z, Xie L, Jiang T, Jiang T J. 2020. Hemolytic and cytotoxic activity from cultures of Aureococcus anophagefferens—a causative species of brown tides in the north-western Bohai Sea, China. Chemosphere, 247: 125819.
DOI:10.1016/j.chemosphere.2020.125819 |
Ji Y Q, Ashton L, Pedley S M, Edwards D P, Tang Y, Nakamura A, Kitching R, Dolman P M, Woodcock P, Edwards F A, Larsen T H, Hsu W W, Benedick S, Hamer K C, Wilcove D S, Bruce C, Wang X Y, Levi T, Lott M, Emerson B C, Yu D W. 2013. Reliable, verifiable and efficient monitoring of biodiversity via metabarcoding. Ecology Letters, 16(10): 1245-1257.
DOI:10.1111/ele.12162 |
Kataoka T, Yamaguchi H, Sato M, Watanabe T, Taniuchi Y, Kuwata A, Kawachi M. 2017. Seasonal and geographical distribution of near-surface small photosynthetic eukaryotes in the western North Pacific determined by pyrosequencing of 18S rDNA. FEMS Microbiology Ecology, 93(2): fiw229.
DOI:10.1093/femsec/fiw229 |
Keeley N, Wood S A, Pochon X. 2018. Development and preliminary validation of a multi-trophic metabarcoding biotic index for monitoring benthic organic enrichment. Ecological Indicators, 85: 1044-1057.
DOI:10.1016/j.ecolind.2017.11.014 |
Kremp A, Elbrachter M, Schweikert M, Wolny J L, Gottschling M. 2005. Woloszynskia halophila (Biecheler) comb. nov. : a bloom-forming cold-water dinoflagellate co-occurring with Scrippsiella hangoei (Dinophyceae) in the Baltic Sea. Journal of Phycology, 41(3): 629-642.
DOI:10.1111/j.1529-8817.2005.00070.x |
Krock B, Tillmann U, Voß D, Koch B P, Salas R, Witt M, Potvin é, Jeong H J. 2012. New azaspiracids in Amphidomataceae (Dinophyceae). Toxicon, 60(5): 830-839.
DOI:10.1016/j.toxicon.2012.05.007 |
Krock B, Tillmann U, Witt M, Gu H F. 2014. Azaspiracid variability of Azadinium poporum (Dinophyceae) from the China Sea. Harmful Algae, 36: 22-28.
DOI:10.1016/j.hal.2014.04.012 |
Lear G, Dickie I, Banks J, Boyer S, Buckley H, Buckley T, Cruickshank R, Dopheide A, Handley K, Hermans S, Kamke J, Lee C, Macdiarmid R, Morales S, Orlovich D, Smissen R, Wood J, Holdaway R. 2018. Methods for the extraction, storage, amplification and sequencing of DNA from environmental samples. New Zealand Journal of Ecology, 42(1): 10.
DOI:10.20417/nzjecol.42.9 |
Lei M D. 2021. Studies on Community Structure of Eukaryotic Algae in Surface Sediments from the Qingdao Coast based on Metabarcoding and its Relationship with Biogenic Elements. Jinan University, Guangzhou, China.
|
LeRoi J M, Hallegraeff G M. 2006. Scale-bearing nanoflagellates from southern Tasmanian coastal waters, Australia. II. Species of Chrysophyceae (Chrysophyta), Prymnesiophyceae (Haptophyta, excluding Chrysochromulina) and Prasinophyceae (Chlorophyta). Botanica Marina, 49(3): 216-235.
DOI:10.1515/BOT.2006.027 |
Li M, Li K Q, Chen K, Liu C C, Ma Y P, Wang X L. 2020. Size-based bioavailability of land-based DON and its impact on eutrophication of Jiaozhou bay. Marine Pollution Bulletin, 152: 110898.
DOI:10.1016/j.marpolbul.2020.110898 |
Li Y, Tang Y N, Shen P P, Gu H F, Tan Y H. 2017. Distribution of dinoflagellate resting cysts in surface sediment of Jiaozhou Bay, China. Oceanologia et Limnologia Sinica, 48(4): 760-766.
(in Chinese with English abstract) |
Limoges A, Mertens K N, Ruíz-Fernández A C, de Vernal A. 2015. First report of fossilized cysts produced by the benthic Bysmatrum subsalsum (Dinophyceae) from a shallow Mexican lagoon in the Gulf of Mexico. Journal of Phycology, 51(1): 211-215.
DOI:10.1016/10.1111/jpy.12257 |
Lin S H, Hu Z X, Deng Y Y, Shang L X, Gobler C J, Tang Y Z. 2020. An assessment on the intrapopulational and intraindividual genetic diversity in LSU rDNA in the harmful algal blooms-forming dinoflagellate Margalefidinium (= Cochlodinium) fulvescens based on clonal cultures and bloom samples from Jiaozhou Bay, China. Harmful Algae, 96: 101821.
DOI:10.1016/j.hal.2020.101821 |
Lin S J. 2011. Genomic understanding of dinoflagellates. Research in Microbiology, 162(6): 551-569.
DOI:10.1016/j.resmic.2011.04.006 |
Liu L, Wang Z H, Lu S H. 2020a. Diversity and geographical distribution of resting stages of eukaryotic algae in the surface sediments from the southern Chinese coastline based on metabarcoding partial 18S rDNA sequences. Marine Ecology, 41(3): 1-17.
DOI:10.1111/maec.12585 |
Liu S M, Zhu B D, Zhang J, Wu Y, Liu G S, Deng B, Zhao M X, Liu G Q, Du J Z, Ren J L, Zhang G L. 2010. Environmental change in Jiaozhou Bay recorded by nutrient components in sediments. Marine Pollution Bulletin, 60(9): 1591-1599.
DOI:10.1016/j.marpolbul.2010.04.003 |
Liu S Y, Chen N S. 2021. Advances in biodiversity analysis of phytoplankton and harmful algal bloom species in the Jiaozhou Bay. Marine Sciences, 45(4): 170-188.
(in Chinese with English abstract) DOI:10.11759/hykx20201021003 |
Liu S Y, Gibson K, Cui Z M, Chen Y, Sun X X, Chen N S. 2020b. Metabarcoding analysis of harmful algal species in Jiaozhou Bay. Harmful Algae, 92: 101772.
DOI:10.1016/j.hal.2020.101772 |
Liu Y Y, Hu Z X, Deng Y Y, Tang Y Z. 2020c. Evidence for resting cyst production in the cosmopolitan toxic dinoflagellate Karlodinium veneficum and the cyst distribution in the China seas. Harmful Algae, 93: 101788.
DOI:10.1016/j.hal.2020.101788 |
Lundholm N, Churro C, Fraga S, Hoppenrath M, Iwataki M, Larsen J, Mertens K, Moestrup Ø, Zingone A. 2009 onwards. IOC-UNESCO Taxonomic Reference List of Harmful Micro Algae. Accessed at https://www.marinespecies.org/hab on 2022-03-06, https://doi.org/10.14284/362.
|
Massana R, Gobet A, Audic S, Bass D, Bittner L, Boutte C, Chambouvet A, Christen R, Claverie J M, Decelle J, Dolan J R, Dunthorn M, Edvardsen B, Forn I, Forster D, Guillou L, Jaillon O, Kooistra W H C F, Logares R, Mahé F, Not F, Ogata H, Pawlowski J, Pernice M C, Probert I, Romac S, Richards T, Santini S, Shalchian-Tabrizi K, Siano R, Simon N, Stoeck T, Vaulot D, Zingone A, de Vargas C. 2015. Marine protist diversity in European coastal waters and sediments as revealed by high-throughput sequencing. Environmental Microbiology, 17(10): 4035-4049.
DOI:10.1111/1462-2920.12955 |
Matsuoka K, Fukuyo Y. 2000. Technical Guide for Modern Dinoflagellate Cyst Study. WESTPACHAB/WESTPAC/IOC, Japan Society of the Promotion Science, Tokyo.
|
McQuoid M R, Hobson L A. 1996. Diatom resting stages. Journal of Phycology, 32(6): 889-902.
DOI:10.1111/j.0022-3646.1996.00889.x |
McQuoid M R. 2002. Pelagic and benthic environmental controls on the spatial distribution of a viable diatom propagule bank on the Swedish west coast. Journal of Phycology, 38(5): 881-893.
DOI:10.1046/j.1529-8817.2002.01169.x |
Mertens K N, Gu H F, Pospelova V, Chomérat N, Nézan E, Gurdebeke P R, Bogus K, Vrielinck H, Rumebe M, Meteigner C. 2017. First record of resting cysts of the benthic dinoflagellate Prorocentrum leve in a natural reservoir in Gujan-Mestras, Gironde, France. Journal of Phycology, 53(6): 1193-1205.
DOI:10.1111/jpy.12582 |
Ministry of Ecology and Environment of the People's Republic of China. 2010-2017. "The China Marine Environmental Quality Bulletin, " (annual report). http://www.mee.gov.cn/hjzl/sthjzk/jagb/. Accessed on 2022-03-06.
|
Moestrup Ø, Lindberg K, Daugbjerg N. 2009. Studies on woloszynskioid dinoflagellates IV: the genus Biecheleria gen.nov.. Phycological Research, 57(3): 203-220.
DOI:10.1111/j.1440-1835.2009.00540.x |
Montresor M, Di Prisco C, Sarno D, Margiotta F, Zingone A. 2013. Diversity and germination patterns of diatom resting stages at a coastal Mediterranean site. Marine Ecology Progress Series, 484: 79-95.
DOI:10.3354/meps10236 |
Mortlock R A, Froelich P N. 1989. A simple method for the rapid determination of biogenic opal in pelagic marine sediments. Deep Sea Research Part A. Oceanographic Research Papers, 36(9): 1415-1426.
DOI:10.1016/0198-0149(89)90092-7 |
Pan H Z, Li A F, Cui Z G, Ding D S, Qu K M, Zheng Y Y, Lu L, Jiang T, Jiang T J. 2020. A comparative study of phytoplankton community structure and biomass determined by HPLC-CHEMTAX and microscopic methods during summer and autumn in the central Bohai Sea, China. Marine Pollution Bulletin, 155: 111172.
DOI:10.1016/j.marpolbul.2020.111172 |
Penna A, Casabianca S, Guerra A F, Vernesi C, Scardi M. 2017. Analysis of phytoplankton assemblage structure in the Mediterranean Sea based on high-throughput sequencing of partial 18S rRNA sequences. Marine Genomics, 36: 49-55.
DOI:10.1016/j.margen.2017.06.001 |
Piredda R, Sarno D, Lange C B, Tomasino M P, Zingone A, Montresor M. 2017. Diatom resting stages in surface sediments: a pilot study comparing next generation sequencing and serial dilution cultures. Cryptogamie, Algologie, 38(1): 31-46.
DOI:10.7872/crya/v38.iss1.2017.31 |
Place A R, Bowers H A, Bachvaroff T R, Adolf J E, Deeds J R, Sheng J. 2012. Karlodinium veneficum-the little dinoflagellate with a big bite. Harmful Algae, 14: 179-195.
DOI:10.1016/j.hal.2011.10.021 |
Pochon X, Wood S A, Keeley N B, Lejzerowicz F, Esling P, Drew J, Pawlowski J. 2015. Accurate assessment of the impact of salmon farming on benthic sediment enrichment using foraminiferal metabarcoding. Marine Pollution Bulletin, 100(1): 370-382.
DOI:10.1016/j.marpolbul.2015.08.022 |
Rhodes L L, Smith K F, MacKenzie L, Moisan C. 2020. Checklist of the planktonic marine dinoflagellates of New Zealand. New Zealand Journal of Marine and Freshwater Research, 54(1): 86-101.
DOI:10.1080/00288330.2019.1626746 |
Rochon A, Lewis J, Ellegaard M, Harding I C. 2009. The Gonyaulax spinifera (Dinophyceae) "complex": perpetuating the paradox?. Review of Palaeobotany and Palynology, 115(1-2): 53-60.
DOI:10.1016/j.revpalbo.2008.12.017 |
Salonen I S, Chronopoulou P M, Leskinen E, Koho K A. 2019. Metabarcoding successfully tracks temporal changes in eukaryotic communities in coastal sediments. FEMS Microbiology Ecology, 95(1): fiy226.
DOI:10.1093/femsec/fiy226 |
Scoble J M, Cavalier-Smith T. 2014. Scale evolution in Paraphysomonadida (Chrysophyceae): sequence phylogeny and revised taxonomy of Paraphysomonas, new genus Clathromonas, and 25 new species. European Journal of Protistology, 50(5): 551-592.
DOI:10.1016/j.ejop.2014.08.001 |
Shang L X, Hu Z X, Deng Y Y, Liu Y Y, Zhai X Y, Chai Z Y, Liu X H, Zhan Z F, Dobbs F C, Tang Y Z. 2019. Metagenomic sequencing identifies highly diverse assemblages of Dinoflagellate cysts in sediments from ships' ballast tanks. Microorganisms, 7(8): 250.
DOI:10.3390/microorganisms7080250 |
Shen P P, Li Y N, Qi Y Z, Zhang L P, Tan Y H, Huang L M. 2012. Morphology and bloom dynamics of Cochlodinium geminatum (Schütt) Schutt in the Pearl River Estuary, South China Sea. Harmful Algae, 13: 10-19.
DOI:10.1016/j.hal.2011.09.009 |
Shi J, Leng Q, Zhu J Y, Gao H W, Guo X Y, Mao X Y. 2020. Influences of nutrient sources on the alternation of nutrient limitations and phytoplankton community in Jiaozhou Bay, southern Yellow Sea of China. Sustainability, 12(6): 2224.
DOI:10.3390/su12062224 |
Smayda T J. 2002. Turbulence, watermass stratification and harmful algal blooms: an alternative view and frontal zones as "pelagic seed banks". Harmful Algae, 1(1): 95-112.
DOI:10.1016/S1568-9883(02)00010-0 |
Song L, Wu J, Du J, Li N, Song G J, Wang K, Sun M, Wang P. 2019. The characteristics and distribution of eukaryotic phytoplankton community in Liaodong Bay, China. Ocean Science Journal, 54(2): 183-203.
DOI:10.1007/s12601-019-0007-9 |
Sundström A M, Kremp A, Tammilehto A, Tuimala J, Larsson U. 2010. Detection of the bloom-forming cold-water dinoflagellate Biecheleria baltica in the Baltic Sea using LSU rRNA probes. Aquatic Microbial Ecology, 61(2): 129-140.
DOI:10.3354/ame01442 |
Thien S J, Myers R. 1992. Determination of bioavailable phosphorus in soil. Soil Science Society of America Journal, 56(3): 814-818.
DOI:10.2136/sssaj1992.03615995005600030023x |
Wang H X, Lu D D, Huang H Y, Göbel J, Dai X F, Xia P. 2011. First observation of Karlodinium veneficum from the East China Sea and the coastal waters of Germany. Acta Oceanologica Sinica, 30(6): 112-121.
DOI:10.1007/s13131-011-0168-6 |
Wang Y Q, Song J M, Duan L Q, Yuan H M, Li X G, Li N, Zhang Q, Liu J. 2021. Historical reconstructions of sedimentary organic matter sources and phytoplankton evolution in the Jiaozhou Bay based on sterols and carbon isotope. Marine Pollution Bulletin, 165: 112109.
DOI:10.1016/j.marpolbul.2021.112109 |
Xu X, Yu Z M, Cheng F J, He L Y, Cao X H, Song X X. 2017. Molecular diversity and ecological characteristics of the eukaryotic phytoplankton community in the coastal waters of the Bohai Sea, China. Harmful Algae, 61: 13-22.
DOI:10.1016/j.hal.2016.11.005 |
Yan T, Zhou M J, Zou J Z. 2002. A national report on harmful algal blooms in China. Harmful Algal Blooms in the PICES Region of the North Pacific. Report 23. https://www.pices.int/publications/scientific_reports/Report23/default.aspx. Accessed 2020-09-11.
|
Yang S M, Wang L S, Shi X Y. 2014. Phytoplankton community of the Jiaozhou Bay in spring 2009. Oceanologia et Limnologia Sinica, 45(6): 1234-1240.
DOI:10.11693/hyhz20140700209 |
Yao P, Yu Z G, Deng C M, Liu S X, Zhen Y. 2010. Spatial temporal distribution of phytoplankton pigments in relation to nutrient status in Jiaozhou Bay, China. Estuarine, Coastal and Shelf Science, 89(3): 234-244.
DOI:10.1016/j.ecss.2010.07.003 |
Yu L, Wu X Q, Zheng X L, Zheng T Y, Xin J, Walther M. 2019. An index system constructed for ecological stress assessment of the coastal zone: a case study of Shandong, China. Journal of Environmental Management, 232: 499-504.
DOI:10.1016/j.jenvman.2018.11.084 |
Yuan Y Q, Yu Z M, Song X X, Cao X H. 2017. Temporal and spatial characteristics of harmful algal blooms in Qingdao Waters, China. Chinese Journal of Oceanology and Limnology, 35(2): 400-414.
DOI:10.1007/s00343-016-5279-7 |
Zhang X D, Kan J J, Wang J, Gu H F, Hu J, Zhao Y, Sun J. 2015. First record of a large-scale bloom-causing species Nannochloropsis granulata (Monodopsidaceae, Eustigmatophyceae) in China Sea waters. Ecotoxicology, 24(7-8): 1430-1441.
DOI:10.1007/s10646-015-1466-0 |
Zhou J, Wang W, Wu Z H, Wang Q X, Wang Y, Gao X. 2020. The basic characteristics and prevention countermeasures of red tide in Shandong coast waters. Marine Environmental Science, 39(4): 537-543.
DOI:10.12111/j.mes.20190043 |
 2022, Vol. 40
2022, Vol. 40


