Institute of Oceanology, Chinese Academy of Sciences
Article Information
- CAI Fangfang, YU Gongliang, LI Renhui
- Description of two new species of Pseudoaliinostoc (Nostocales, Cyanobacteria) from China based on the polyphasic approach
- Journal of Oceanology and Limnology, 40(3): 1233-1244
- http://dx.doi.org/10.1007/s00343-021-1111-0
Article History
- Received Mar. 31, 2021
- accepted in principle May. 27, 2021
- accepted for publication Jul. 2, 2021
2 Key Laboratory of Algal Biology, State Key Laboratory of Freshwater Ecology and Biotechnology of China, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan 430072, China;
3 School of Life and Environmental Sciences, Wenzhou University, Wenzhou 325035, China
Since cyanobacteria are diverse in morphology, the traditionally classification of cyanobacteria was mainly based on morphological characteristics for a long time (Komárek, 2003; Taton et al., 2003, 2006; Johansen and Casamatta, 2005; Turicchia et al., 2009; Genuário et al., 2013, and many others), but morphology alone seems to be an inadequate tool in the modern taxonomy. With the progress and development of modern science, the introduction of modern methods such as molecular biology and electron microscopy is necessary for taxonomic classification of cyanobacteria (Komárek, 2016). In recent years, with the application of polyphasic approach to cyanobacteria, the taxonomy of cyanobacteria has undergone extensive reconstruction and revision (Komárek et al., 2014). Cyanobacteria currently include eight orders: Gloeobacteriales, Spirulinales, Synechococcales, Chroococcales, Oscillatoriales, Pleurocapsales, Chroococcidiopsidales, and Nostocales (Komárek et al., 2014). Since 2000, more than 140 new genera have been defined, mainly based on genetic criteria. However, there still exist several unsolved problems in this newly introduced method (Komárek, 2020). Under the circumstances, Komárek (2020) demonstrated that modern classification must be applied consistently the polyphasic approach based on phylogenetic classification, but it is necessary to combine and add all cytomorphological and ecological important data, and proper use of nomenclature prescriptions.
The genus Nostoc in the family Nostocaceae has repeatedly been shown to be genetically heterogeneous and polyphyletic (Hrouzek et al., 2005, 2013; Rajaniemi et al., 2005; Papaefthimiou et al., 2008; Lukešová et al., 2009; Silva et al., 2014). Several Nostoc-like genera have been split out from 'Nostoc sensu stricto' clade in an attempt to achieve monophyly in this genus, including Mojavia (Řeháková et al., 2007), Desmonostoc (Hrouzek et al., 2013), Halotia (Genuário et al., 2015), Aliinostoc (Bagchi et al., 2017), Komarekiella (Hentschke et al., 2017), Desikacharya (Saraf et al., 2019), Minunostoc (Cai et al., 2019a), Compactonostoc (Cai et al., 2019b), Purpureonostoc (Cai and Li, 2020; Cai et al., 2020b), and Violetonostoc (Cai et al., 2020a).
The genus Aliinostoc Bagchi, Dubey & Singh (Bagchi et al., 2017) was one example separating from Nostoc, morphologically similar to Nostoc, while the phylogenetic location was in a distant group. The species Aliinostoc morphoplasticum, as the type species, was isolated from eutrophic pond in India. Up to now, six species, A. morphoplasticum, A. catenatum, A. constrictum, A. magnakinetifex, A. soli, and A. tiwarii, have been reported under this genus. However, Cai et al. (2020b) have found that the definition of Aliinostoc was too broad, and this genus has been shown to be polyphyletic in their analyses, resulting that some Aliinostoc species need to be revisited. In accordance with Cai et al. (2020b), Lee et al. (2021) further verified this suggestion, and the members of the genus Aliinostoc have been clustered into two phylogenetically distant nodes. Base on their observation, Lee et al. (2021) convinced that the studied Korean Aliinostoc-like strains, including A. constrictum, A. soli, and A. tiwarii, should be separated into a new genus. As a consequence, Pseudoaliinostoc gen. nov. has been described from the separation of Aliinostoc, with P. soli comb. nov. as the type species. Four Pseudoaliinostoc species, namely, type species P. soli, P. constrictum, P. tiwarii, and P. sejongens have been taxonomically evaluated and described at present.
In the present study, we isolated two Nostoc-like strains from two different sampling areas in China, such as moist soil in Jiangxi province (CHAB5870) and water sample in Dali city, Yunnan province (CHAB5871). Since it is very difficult to differentiate the Nostoc-like group only according to morphological characteristics, phylogenetic studies must therefore be carried out to confirm the correct taxonomic status of all strains. A polyphasic approach was employed to intensively study these two strains. Morphologically, we concluded that both are Nostoc-related morphotypes and the molecular analyses confirmed their position in the Pseudoaliinostoc clade. Our aim in this study is to present the phylogenetic position of these two strains, distinguish them from other Pseudoaliinostoc species and demonstrate that they belong to novel species. Pseudoaliinostoc jiangxiense sp. nov. and Pseudoaliinostoc yunnanense sp. nov. are here taxonomically described, in accordance with the International Code of nomenclature for algae, fungi, and plants.
2 MATERIAL AND METHOD 2.1 Sampling, isolation, and culturing of strainsThe cyanobacterial samples presented in this study were collected from two different localities of China. The strain CHAB5870 was collected from moist soil in Jiangxi Province, China (25°85.09'N, 114°94.02'E), on March 25, 2017. The strain CHAB5871 was isolated from a water sample in Dali city, Yunnan Province, China (26°11.11'N, 99°95.08'E), on August 6, 2016. Under microscope (Olympus C31, Japan), single filaments from the cyanobacterial samples were separated by using lab-made pasteur pipette washing method and then cultured in spiral cap tubes containing 5 mL of CT medium (Watanabe and Ichimura, 1977). All individual isolates were subsequently maintained at 25 ℃, 12 h꞉12 h in a light-dark cycle with a photon flux density of 40 μmol/(m2·s) under white fluorescent light. The living cyanobacterial strains were cultured in the Chinese Harmful Algae Biology (CHAB) culture collection of the Institute of Hydrobiology (IHB), Chinese Academy of Sciences (CAS). Dry materials of strains were freeze-dried at -50 ℃ and stored in the Freshwater Algal Herbarium (HBI), IHB, CAS.
2.2 Morphological characterizationMorphological characterization of the strains CHAB5870 and CHAB5781 was performed using Nikon Eclipse 80i microscope (Nikon, Tokyo, Japan) using its differential interference contrast microscopy. The micrographs were taking using Nikon software NIS-Elements 3.2D (Nikon, Tokyo, Japan). The Nikon Eclipse 80i microscope equipped with a Nikon DS-Ri1 digital camera was used to observed and described the shape and sizes of vegetative cells, heterocytes, and akinetes (n≥50), as well as the presence or absence of sheaths. The sizes are presented by "min-average-max".
2.3 DNA extraction and polymerase chain reactionTotal genomic DNA was isolated from 25-day old liquid cultures of unialgal cyanobacterial strains using Clarke's method (Clarke, 2009). Amplification of 16S rRNA gene was performed using primer pA (5'-AGAGTTTGATCCTGGCTCAG-3') and primer B23S (5'-CTTCGCCTCTGTGTGCCTAGGT-3') (Edwards et al., 1989; Gkelis et al., 2005). Primers 322 and 340 (Iteman et al., 2000) were used for amplification of the 16S-23S ITS region. The PCR reaction was carried out in 20-μL reaction mixture containing 1 μL of genomic DNA (100 ng/μL), 0.5 μL of each primer (10 μmol/L), 10 μL of 2× PCR mix with Taq polymerase (Cat TSE001, Beijing Tsingke Biotech Co. Ltd., Beijing, China), and 8 μL of sterile water. The PCR was conducted on an MJ Mini Personal Thermal Cycler (Bio-Rad, Hercules, California USA), and the PCR cycle had initial denaturation at 95 ℃ for 3 min, followed by 33 cycles of 95 ℃ for 30 s, 58 ℃ for 30 s (30 s at 55 ℃ for ITS), 72 ℃ for 1 min (30 s for ITS), and a final 5-min elongation step at 72 ℃. The resulting PCR products were purified using TSINGKE DNA Gel Extraction Kit (Cat GE0101-200, Beijing Tsingke Biotech Co. Ltd., Beijing, China) and subsequently cloned into pMDTM18-T vector (TaKaRa, Japan) using the procedure of Sambrook and Russell (2001). All sequencing was performed by the ABI 3730 Automated Sequencer (PerkinElmer, Waltham, MA, USA). The 16S rRNA and 16S-23S ITS gene sequences of the cyanobacterial strains CHAB5870 and CHAB5871 were deposited in the NCBI (National Center for Biotechnology Information, https://www.ncbi.nlm.nih.gov/) GenBank database under accession numbers: MW829373, MW829374, MW829375, MW829376, and MW829377.
2.4 Phylogenetic analysis16S rRNA gene sequences obtained in the present study and those representing main groups of heterocytous cyanobacteria retrieved from GenBank were used for phylogenetic analyses. All downloaded sequences were edited by using MAFFT v7.312 (Katoh and Standley, 2013) with auto-selected strategy FFT-NS-I (with default parameters) after multiple sequence alignment, and visually checked in MEGA v.7.0.14 (Kumar et al., 2016). The 16S rRNA gene phylogenetic trees were inferred by maximum-parismony (MP), maximum likelihood (ML), and bayesian inference (BI) methods. The MP analysis with 1 000 repeated heuristic searches was performed by MEGA software X (Kumar et al., 2018). The best-fit models, GTR+I+G, were selected for the ML, MP, and BI analysis under the Akaike Information Criterion (AIC) in ModelFinder (Kalyaanamoorthy et al., 2017). The BI was calculated with MrBayes v3.2.6 (Ronquist et al., 2012) in the CIPRES Science Gateway V.3.3 (Miller et al., 2015, http://www.phylo.org/). In the BI analyses, two runs of eight Markov chains were run for 8 million generations, sampling every 100 generations, with 25% of the sampled trees discarded as burn-in, the final average standard deviation of splitting frequency was < 0.01. In the ML analyses, 10 000 bootstrap replicates were performed to evaluate the relative support of the branches by using ultrafast bootstrap on IQ-TREE web server (Trifinopoulos et al., 2016). The obtained consensus phylogenetic trees were visualized in FigTree, v1.4.3 (Rambaut, 2016) with Chroococcidiopsis thermalis PCC7203 as the outgroup. The p-distance was calculated with pairwise deletion of gaps by MEGA software v.7.0.14 (Kumar et al., 2016) and used to calculate sequence identity [100×(1-p)] for 16S rRNA data.
2.5 Analyses of 16S-23S internal transcribed spacer (ITS)The ITS secondary structures of D1-D1' and Box-B helix of the two studied strains and other closely species were determined using software RNAstructure ver. 5.6 (Mathews Lab, 2013).
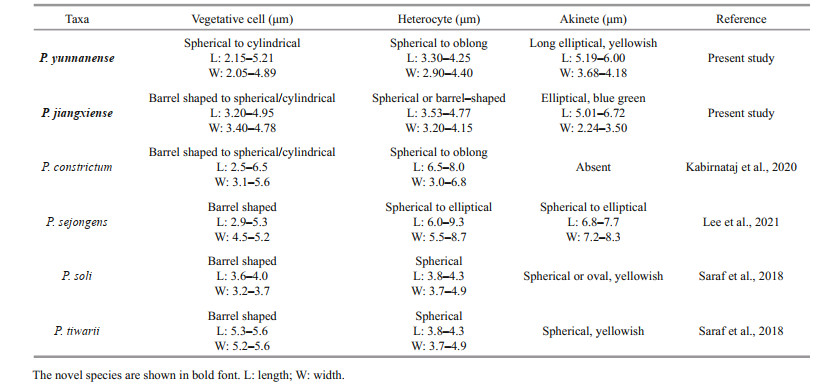
3 RESULT 3.1 MorphologyPseudoaliinostoc jiangxiense F. Cai et R. Li sp. nov. (Fig. 1)
|
| Fig.1 Micrographs of Pseudoaliinostoc jiangxiense under the light microscopy (LM) a. general view of microcolonies; b-d. single trichomes; e-h. filaments with heterocytes and akinetes. H: heterocytes; ak: akinetes, sh: sheath. Scale bars: 10 μm. |
Description: Growing as a soft macroscopic mat on wet soils, with visible blue green color. Filaments loosely or densely entangled, sometimes straight. Sheath colorless, thin, or slightly thick, distinct or gelatinized. Trichomes isopolar, straightly or curved, blue-green, distinctly constricted at the cross-walls, attenuated at the ends. Vegetative cells barrel-shaped to spherical or cylindrical, 3.20-3.41-4.95-μm long, 3.40-3.62-4.78-μm wide. Heterocytes usually intercalary, rarely terminal, spherical or barrel-shaped, 3.53-3.84-4.77-μm long, 3.20-3.59-4.15-μm wide. Akinetes intercalary, elliptical, 5.01-5.85-6.72-μm long, 2.24-3.24-3.50-μm wide.
Holotype here designated: Dry material of the strain CHAB5870 was stored at the Freshwater Algal Herbarium (HBI), Institute of Hydrobiology, Chinese Academy of Science, Wuhan, China, as specimen No. JXGJ201701.
Habitat: wet soils.
Type locality: Isolated from wet soils in Jiangxi province, China (25°85.09′N, 114°94.02′E).
Reference strain: The living culture was deposited in Collection of Harmful Algae Biology (CHAB), Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, Hubei Province, China as strain CHAB5870.
Etymology: jiangxiense refers to Jiangxi province where the strain was isolated, transliterated into Latin.
Pseudoaliinostoc yunnanense F. Cai et R. Li sp. nov. (Fig. 2)

|
| Fig.2 Micrographs of Pseudoaliinostoc yunnanense under the light microscopy (LM) a. filaments with akinetes; b, c, e, f. single trichomes; d. filamentous with internal heterocytes and terminal heterocytes. H: heterocytes; ak: akinetes. Scale bars: 10 μm. |
Description: Filaments straight, loosely or densely entangled. Sheath colorless, thin, usually invisible. Trichomes isopolar, straightly or curved, blue-green, distinctly constricted at the cross-walls. Vegetative cells spherical to cylindrical, 2.15-3.79-5.21-μm long, 2.05-2.25-4.89-μm wide. Heterocytes usually intercalary, rarely terminal, 3.30-3.59-4.25-μm long, 2.90-3.05-4.40-μm wide. Akinetes intercalary, oval, 5.19-5.72-6.00-μm long, 3.68-3.87-4.18-μm wide.
Holotype here designated: Dry material of the strain CHAB5871 was stored at the Freshwater Algal Herbarium (HBI), Institute of Hydrobiology, Chinese Academy of Science, Wuhan, China, as specimen No. YNDL201601.
Habitat: Free-living in water.
Type locality: Isolated from a water sample in Yunnan Province, China (26°11.11'N, 99°95.08'E).
Reference strains: The living culture was deposited in Collection of Harmful Algae Biology (CHAB), Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, Hubei Province, China as strain CHAB5871.
Etymology: yunnanense refers to Yunnan province where the strain was isolated, transliterated into Latin.
3.2 Molecular and phylogenetic analysesThe 16S rRNA gene sequences of the studied strains were obtained and evaluated with BLAST analyses in NCBI. The 16S rRNA gene sequence of the strain CHAB5870 showed 93.7%-94.8% similarity with existing Aliinostoc species, 96.3%- 98.0% similarity with existing Pseudoaliinostoc species, and 94.2% similarity with Purpureonostoc species. The strain CHAB5871 containing two clones shared 99.7% similarity with each other, 91.9%- 93.3% similarity with Aliinostoc species, 95.5%-96.6% similarity with Pseudoaliinostoc species, and 92.1% similarity with Purpureonostoc species. The sequence similarity of the two strains described in this study were also compared and presented with percent identity of 95.8% (Table 1).

|
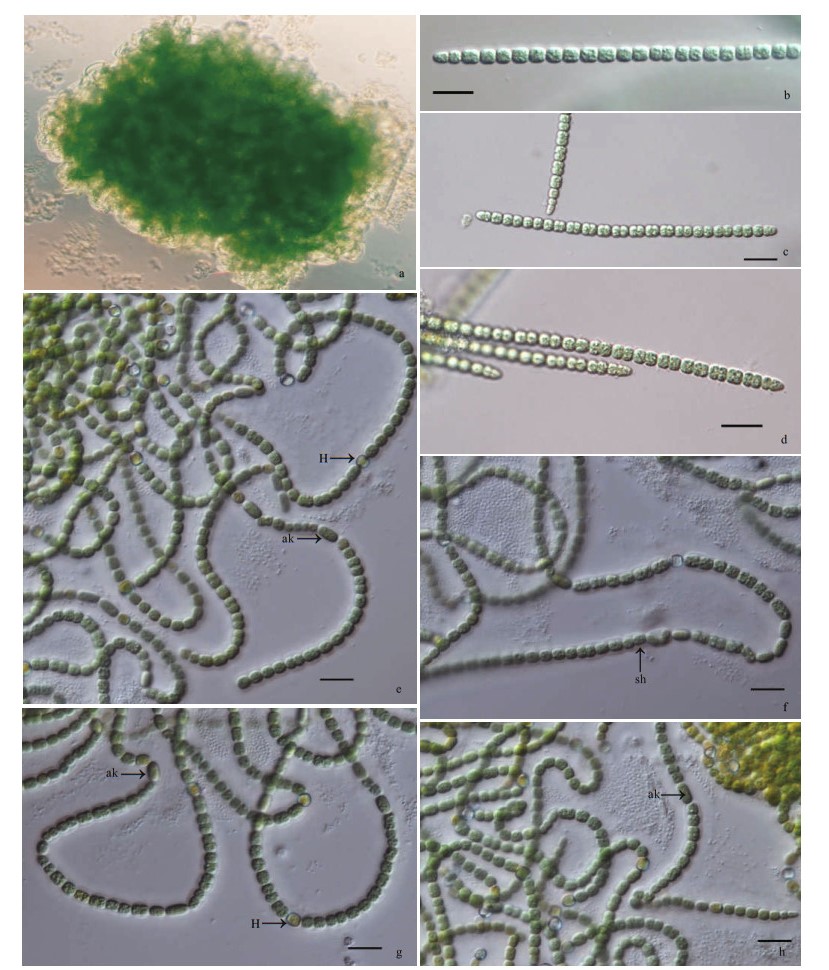
Accession numbers for GenBank sequences that were used for phylogenetic analyses were listed as a supplementary file (Supplementary Table S1). The16S rRNA gene phylogenetic trees, from 164 cyanobacterial groups including the studied strains, were constructed by the MP, ML, and BI methods. The Bayesian tree revealed that the studied Pseudoaliinostoc strains, together with other Pseudoaliinostoc species, formed a single and unique cluster, and this distinctive clade was supported by MP, ML, and BI approaches with high bootstrap values of 100%, 100%, and 1.00, respectively (Fig. 3).
|
| Fig.3 Bayesian inference (BI) phylogenetic tree based on 16S rDNA sequences (1 157 bp) of the studied strains and other cyanobacterial strains Bootstrap values greater than 50% with MP/ML/BI methods are indicated on the tree, and the asterisks at the nodes mean 100. The novel species is in bold font. |
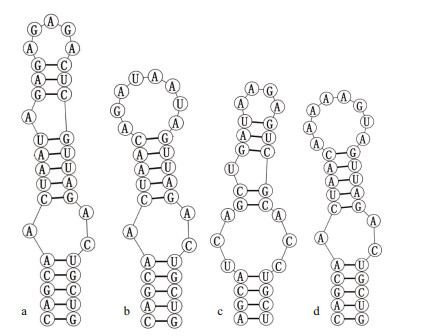
The ITS secondary structure of D1-D1' of the strains CHAB5870 and CHAB5871 were compared amongst themselves and with the four existing Pseudoaliinostoc species. For Box-B helix, the secondary structures were obtained for two studied strains, P. constrictum SA30 and P. sejongens FBCC-A406, but except for P. soli ZH1(3)_PS and P. tiwarii Li_PS. Several species in this genus lack V3 helix (Saraf et al., 2018; Kabirnataj et al., 2020), so this structure has not been presented in the present study.
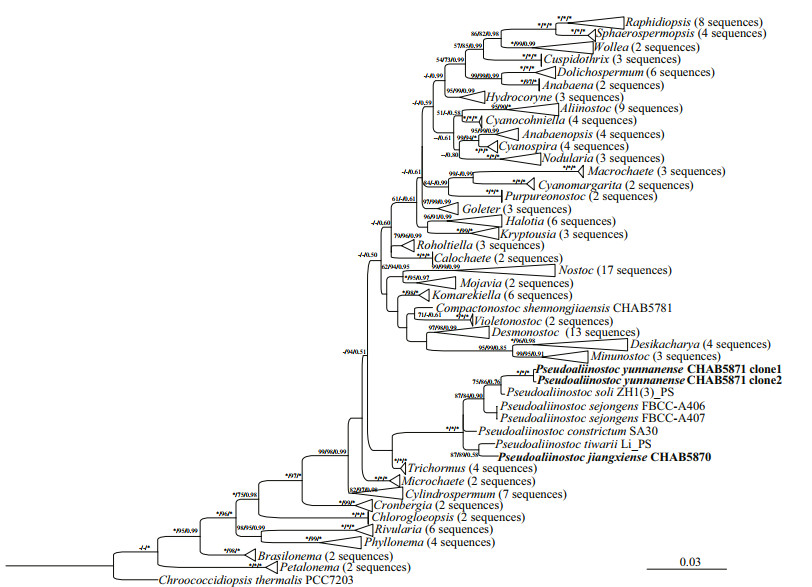
Analyses of D1-D1' helix (Fig. 4) of the two studied strains showed unique structure and different nucleotides in comparison with other Pseudoaliinostoc species. Six Pseudoaliinostoc strains were divided into six types. The D1-D1' helix structure of the strain CHAB5870 showed that the basal stem consisted of a 6-bp helix, followed by a 1꞉7-bp bilateral bulge, a 1꞉1-base bilateral bulge (A꞉A), the bilateral bulge was connected by a side loop with a single base on the 5ʹ side, following this side loop was one bilateral bulge of 4꞉5-bases, the terminal loop contained 5-bp bases. The base stem of the D1-D1' helix of the strain CHAB5871 consisted of 5-bp helix, followed by a big side loop with 7-bp bases on the 3ʹ side (other closely species do not possess this unique structure), and then further followed by three bilateral bulges of 3꞉2-, 2꞉1-, and 4꞉5-bases, the terminal loop contained 5-bp bases. Shown in Fig. 5, the Box-B helix of the strains CHAB5870 and CHAB5871 presented unique structure when compared with other taxa. The Box-B helix of the strain CHAB5870 was comprised of a 5-bp helix, followed by a 1꞉2-bp bilateral bulge, an 8-bp stem with on side loop with a single base on the 5ʹ side, and the terminal loop with 5 bases (AGAGA). The strain CHAB5871 consisted of 5-bp helix in the base of the stem, followed by a 1꞉2 base bilateral bulge, the terminal loop contained 8-bp bases (AGAUAAUA).
|
| Fig.4 Secondary structure of D1-D1ʹ helix a. Pseudoaliinostoc jiangxiense CHAB5870; b. Pseudoaliinostoc yunnanense CHAB5871; c. Pseudoaliinostoc soli ZH1(3)_PS; d. Pseudoaliinostoc tiwarii Li_PS; e. Pseudoaliinostoc constrictum SA30; f. Pseudoaliinostoc sejongens FBCC-A406. |
|
| Fig.5 Secondary structure of Box-B helix a. Pseudoaliinostoc jiangxiense CHAB5870; b. Pseudoaliinostoc yunnanense CHAB5871; c. Pseudoaliinostoc constrictum SA30; d. Pseudoaliinostoc sejongens FBCC-A406. |
In recent years, with the development of phylogenetic analysis methods based on molecular sequence data, the classification system of cyanobacteria has been extensively revised and reconstructed. It is universally recognized that ideal genera and species of cyanobacteria should be monophyletic in the modern cyanobacterial taxonomy (Komárek et al., 2014; Miscoe et al., 2016; Komárek, 2018). As one of the most problematic taxonomic group, Family Nostocaceae with more or less complicated filaments has received significantly studies, and several genera of this family have been shown to be polyphyletic (Rajaniemi et al., 2005; Kozhevnikov and Kozhevnikova, 2011; Zapomělová et al., 2013; Komárek et al., 2014; Choi et al., 2018). Recent efforts have been made to reassess the genus Nostoc (the type genus of the family Nostocaceae) by identifying Nostoc sensu stricto, a clade including the type species Nostoc commune, and excluding the species that fall outside of this clade with Nostoc name, but cannot be classified into that genus if monophyly must be achieved at the genus level. Building these novel genera Mojavia (Řeháková et al., 2007), Desmonostoc (Hrouzek et al., 2013), Halotia (Genuário et al., 2015), Aliinostoc (Bagchi et al., 2017), Komarekiella (Hentschke et al., 2017) and Desikacharya (Saraf et al., 2019), Minunostoc (Cai et al., 2019a), Compactonostoc (Cai et al., 2019b), Purpureonostoc (Cai and Li, 2020; Cai et al., 2020b), and Violetonostoc (Cai et al., 2020a) by separating from the Nostoc sensu stricto clade was significant example representing the effort to make Nostoc monophyletic.
The newly described genus Aliinostoc, with six species at present, has been shown to be genetically heterogeneous and polyphyletic, and members of this genus have been divided into two distinct clades (Cai et al., 2020b; Lee et al., 2021). In this case, A. constrictum, A. soli, and A. tiwarii has been separated from Aliinostoc to form a new genus Pseudoaliinostoc (Lee et al., 2021) in an attempt to make Aliinostoc monophyly. In this study, two strains CHAB5870 and CHAB5871 isolated from two different ecology were characterized by the polyphasic approach. The preliminary morphological characteristics of the strains indicated that they were members of the complex Nostoc-like group. In the 16S rRNA gene phylogenetic tree, the strains CHAB5870 and CHAB5871, along with Pseudoaliinostoc soli ZH1(3)_PS, Pseudoaliinostoc tiwarii Li_PS, Pseudoaliinostoc constrictum SA30, and Pseudoaliinostoc sejongens FBCC-A406 formed a distinct clade, namely the newly described Pseudoaliinostoc clade. In addition, the strong bootstrap support for the whole Pseudoaliinostoc clade indicated that the strains CHAB5870 and CHAB5871 were members of the genus Pseudoaliinostoc.
Based on the evolutionary distance matrix, the 16S rRNA gene sequences of the strains CHAB5870 and CHAB5871 showed ≤98% similarity with existing Pseudoaliinostoc species. In bacteriology, it has been recommended by some researchers to use percentage identities of less than 97.5% 16S rRNA gene sequences to distinguish species (Stackebrandt and Goebel, 1994), however, based on extensive comparison of more strains, Stackebrandt and Ebers (2006) recommended 98.7%-99.0% of the 16S rRNA gene sequence similarity threshold as species separation. Other research suggested that through twofold cross-validation statistical test, 98.65% of the 16S rRNA gene sequence similarity could be a threshold to differentiate two species (Kim et al., 2014). Therefore, in combination with the 16S rRNA gene sequence similarity value recently proposed, the strains CHAB5870 and CHAB5871 shared ≤98% similarity with the existing Pseudoaliinostoc species, indicating that they are new members of the genus Pseudoaliinostoc.
The analysis of 16S-23S internal transcribed spacer (ITS), such as secondary structure, is an effective tool for species separation and has been demonstrated in a plenty of studies (Johansen et al., 2011; Vaccarino and Johansen, 2012; Osorio-Santos et al., 2014; Bohunická et al., 2015; Sciuto and Moro, 2016; Mai et al., 2018, and many others). In this study, the ITS secondary structures of D1-D1ʹ and Box-B helix of the two strains were quite different investigated in comparison with other Pseudoaliinostoc species, which enabled our strains to be distinguished from other Pseudoaliinostoc species.
The morphological characters of the two studied strains were also compared with the species of Pseudoaliinostoc (Table 2). The strain CHAB 5870 differs from P. sejongens in having narrower akinetes and heterocytes. Vegetative cells in CHAB5870 have smaller maximum dimensions than cells in P. tiwarii. CHAB5870 and P. soli differs in the shape and color of the akinetes. Compared to P. tiwarii, CHAB5871 has smaller width vegetative cells. The akinetes and heterocytes of P. sejongens have width lager than CHAB5871. CHAB5871 and P. soli differs in the shape and size of the akinetes. The difference between the two studied strains in this study is that CHAB5871 has wider akinetes than CHAB5870. The morphological difference among P. constrictum, CHAB5870 and CHAB5871 was whether akinete could be observed present or absent: present in CHAB5870 and CHAB5871 and absent in P. constrictum. Minor morphological differences were observed, which also indicated the two studied strains to be novel species of the genus Pseudoaliinostoc.
By comparing polyphasic taxonomy of two Nostoc-like strains, i.e., CHAB5870 and CHAB5871, and analyzing their morphology, 16S rRNA phylogeny, and ITS secondary structures, we strongly believed that the strains represent two novel species of the genus Pseudoaliinostoc: Pseudoaliinostocjiangxiense sp. nov. and Pseudoaliinostoc yunnanense sp. nov., for which morphological descriptions are given. This is the first report of the two new Pseudoaliinostoc species from China. This report is helpful to expand the knowledge about the species diversity and geographical distribution of the Nostoc-like cyanobacteria in China.
6 DATA AVAILABILITY STATEMENTThe data that support the findings of the study are available from the corresponding author upon reasonable request.
Electronic supplementary materialSupplementary material (Supplementary Table S1) is available in the online version of this article at https://doi.org/10.1007/s00343-021-1111-0.
Bagchi S N, Dubey N, Singh P. 2017. Phylogenetically distant clade of Nostoc-like taxa with the description of Aliinostoc gen. nov. and Aliinostoc morphoplasticum sp. nov. International Journal of Systematic and Evolutionary Microbiology, 67(9): 3329-3338.
DOI:10.1099/ijsem.0.002112 |
Bohunická M, Pietrasiak N, Johansen J R, Gómez E B, Hauer T, Gaysina L A, Lukešová A. 2015. Roholtiella, gen. nov. (Nostocales, Cyanobacteria)—a tapering and branching cyanobacteria of the family Nostocaceae. Phytotaxa, 197(2): 84-103.
DOI:10.11646/phytotaxa.197.2.2 |
Cai F F, Li R H. 2020. Purpureonostoc, a new name for a recently described genus of Nostoc-like cyanobacteria. Fottea, 20(2): 111.
DOI:10.5507/fot.2020.007 |
Cai F F, Li X C, Geng R Z, Peng X, Li R H. 2019a. Phylogenetically distant clade of Nostoc-like taxa with the description of Minunostoc gen. nov. and Minunostoc cylindricum sp. nov. Fottea, 19(1): 13-24.
DOI:10.5507/fot.2018.013 |
Cai F F, Li X C, Yang Y M, Jia N N, Huo D, Li R H. 2019b. Compactonostoc shennongjiaensis gen. & sp. nov. (Nostocales, Cyanobacteria) from a wet rocky wall in China. Phycologia, 58(2): 200-210.
DOI:10.1080/00318884.2018.1541270 |
Cai F F, Peng X, Li R H. 2020a. Violetonostoc minutum gen. et sp. nov. (Nostocales, Cyanobacteria) from a rocky substrate in China. Algae, 35(1): 1-15.
DOI:10.4490/algae.2020.35.3.4 |
Cai F F, Wang Y L, Yu G L, Wang J, Peng X, Li R H. 2020b. Proposal of Purpurea gen. nov. (Nostocales, cyanobacteria), a novel cyanobacterial genus from wet soil samples in Tibet, China. Fottea, 20(1): 86-97.
DOI:10.5507/fot.2019.018 |
Choi H J, Joo J H, Kim J H, Wang P B, Ki J S, Han M S. 2018. Morphological characterization and molecular phylogenetic analysis of Dolichospermum hangangense (Nostocales, Cyanobacteria) sp. nov. from Han River, Korea. Algae, 33(2): 143-156.
DOI:10.4490/algae.2018.33.5.2 |
Clarke J D. 2009. Cetyltrimethyl ammonium bromide (CTAB) DNA miniprep for plant DNA isolation. Cold Spring Harbor Protocols, 2009(3): pdb. prot5177.
DOI:10.1101/pdb.prot5177 |
Edwards U, Rogall T, Blöcker H, Emde M, Böttger E C. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Research, 17(19): 7843-7853.
DOI:10.1093/nar/17.19.7843 |
Genuário D B, Corrêa D M, Komárek J, Fiore M F. 2013. Characterization of freshwater benthic biofilm-forming Hydrocoryne (Cyanobacteria) isolates from Antarctica. Journal of Phycology, 49(6): 1142-1153.
DOI:10.1111/jpy.12124 |
Genuário D B, Vaz M G M V, Hentschke G S, Sant'Anna C L, Fiore M F. 2015. Halotia gen. nov., a phylogenetically and physiologically coherent cyanobacterial genus isolated from marine coastal environments. International Journal of Systematic and Evolutionary Microbiology, 65(Pt_2): 663-675.
DOI:10.1099/ijs.0.070078-0 |
Gkelis S, Rajaniemi P, Vardaka E, Moustaka-Gouni M, Lanaras T, Sivonen K. 2005. Limnothrix redekei (Van Goor) Meffert (Cyanobacteria) strains from Lake Kastoria, Greece form a separate phylogenetic group. Microbial Ecology, 49(1): 176-182.
DOI:10.1007/s00248-003-2030-7 |
Hentschke G S, Johansen J R, Pietrasiak N, Rigonato J, Fiore M F, Sant'anna C L. 2017. Komarekiella atlantica gen. et sp. nov. (Nostocaceae, Cyanobacteria): a new subaerial taxon from the Atlantic Rainforest and Kauai, Hawaii. Fottea, 17(2): 178-190.
DOI:10.5507/fot.2017.002 |
Hrouzek P, Lukešová A, Mareš J, Ventura S. 2013. Description of the cyanobacterial genus Desmonostoc gen. nov. including D. muscorum comb. nov. as a distinct, phylogenetically coherent taxon related to the genus Nostoc. Fottea, 13(2): 201-213.
DOI:10.5507/fot.2013.016 |
Hrouzek P, Ventura S, Lukešová A, Mugnai M, Angela Turicchia S, Komárek J. 2005. Diversity of soil Nostoc strains: phylogenetic and phenotypic variability. Algological Studies, (117): 251-264.
DOI:10.1127/1864-1318/2005/0117-0251 |
Iteman I, Rippka R, De Marsac N T, Herdman M. 2000. Comparison of conserved structural and regulatory domains within divergent 16S rRNA-23S rRNA spacer sequences of cyanobacteria. Microbiology, 146(6): 1275-1286.
DOI:10.1099/00221287-146-6-1275 |
Johansen J R, Casamatta D A. 2005. Recognizing cyanobacterial diversity through adoption of a new species paradigm. Algological Studies, (117): 71-93.
DOI:10.1127/1864-1318/2005/0117-0071 |
Johansen J R, Kovacik L, Casamatta D A, Iková K F, Kaštovský J. 2011. Utility of 16S-23S ITS sequence and secondary structure for recognition of intrageneric and intergeneric limits within cyanobacterial taxa: Leptolyngbya corticola sp. nov. (Pseudanabaenaceae, Cyanobacteria). Nova Hedwigia, 92(3-4): 283-302.
DOI:10.1127/0029-5035/2011/0092-0283 |
Kabirnataj S, Nematzadeh G A, Talebi A F, Saraf A, Suradkar A, Tabatabaei M, Singh P. 2020. Description of novel species of Aliinostoc, Desikacharya and Desmonostoc using a polyphasic approach. International Journal of Systematic and Evolutionary Microbiology, 70(5): 3413-3426.
DOI:10.1099/ijsem.0.004188 |
Kalyaanamoorthy S, Minh B Q, Wong T K F, Von Haeseler A, Jermiin L S. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods, 14(6): 587-589.
DOI:10.1038/nmeth.4285 |
Katoh K, Standley D M. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution, 30(4): 772-780.
DOI:10.1093/molbev/mst010 |
Kim M, Oh H S, Park S C, Chun J. 2014. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. International Journal of Systematic and Evolutionary Microbiology, 64(Pt_2): 346-351.
DOI:10.1099/ijs.0.059774-0 |
Komárek J, Kaštovský J, Mareš J, Johansen J R. 2014. Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) 2014, using a polyphasic approach. Preslia, 86(4): 295-335.
|
Komárek J. 2003. Problem of the taxonomic category "species" in cyanobacteria. Algological Studies, (109): 281-297.
DOI:10.1127/1864-1318/2003/0109-0281 |
Komárek J. 2016. A polyphasic approach for the taxonomy of cyanobacteria: principles and applications. European Journal of Phycology, 51(3): 346-353.
DOI:10.1080/09670262.2016.1163738 |
Komárek J. 2018. Several problems of the polyphasic approach in the modern cyanobacterial system. Hydrobiologia, 811(1): 7-17.
DOI:10.1007/s10750-017-3379-9 |
Komárek J. 2020. Quo vadis, taxonomy of cyanobacteria (2019). Fottea, 20(1): 104-110.
DOI:10.5507/fot.2019.020 |
Kozhevnikov I V, Kozhevnikova N A. 2011. Phylogenetic and morphological evaluation of Wollea saccata (Nostocales, Cyanobacteria) isolated from the Yenissei River basin (Eastern Siberia, Russia). Fottea, 11(1): 99-106.
DOI:10.5507/fot.2011.010 |
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35(6): 1547-1549.
DOI:10.1093/molbev/msy096 |
Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33(7): 1870-1874.
DOI:10.1093/molbev/msw054 |
Lee N J, Bang S D, Kim T, Ki J S, Lee O M. 2021. Pseudoaliinostoc sejongens gen. & sp. nov. (Nostocales, Cyanobacteria) from floodplain soil of the Geum River in Korea based on polyphasic approach. Phytotaxa, 479(1): 55-70.
DOI:10.11646/phytotaxa.479.1.4 |
Lukešová A, Johansen J R, Martin M P, Casamatta D A. 2009. Aulosira bohemensis sp. nov. : further phylogenetic uncertainty at the base of the Nostocales (Cyanobacteria). Phycologia, 48(2): 118-129.
DOI:10.2216/08-56.1 |
Mai T, Johansen J R, Pietrasiak N, Bohunická M, Martin M P. 2018. Revision of the Synechococcales (cyanobacteria) through recognition of four families including Oculatellaceae fam. nov. and six new genera containing 14 species. Phytotaxa, 365(1): 1-59.
DOI:10.11646/phytotaxa.365.1.1 |
Mathews Lab. 2021. RNAstructure, version 6.3. http://rna.urmc.rochester.edu/RNAstructure.html.
|
Miller M A, Schwartz T, Pickett B, He S, Klem E B, Scheuermann R H, Passarotti M, Kaufman S, O'Leary M A. 2015. A RESTful API for access to phylogenetic tools via the CIPRES science gateway. Evolutionary Bioinformatics, 11: 43-48.
DOI:10.4137/EBO.S21501 |
Miscoe L H, Johansen J R, Vaccarino M A, Pietrasiak N, Sherwood A R. 2016. Novel cyanobacteria from caves on Kauai, Hawaii. In: Miscoe L H ed. The Diatom Flora and Cyanobacteria from Caves on Kauai, Hawaii: Taxonomy, Distribution, New Species. Bibliotheca Phycologica, Stuttgart. p. 75-152.
|
Osorio-Santos K, Pietrasiak N, Bohunická M, Miscoe L H, Kováčik L, Martin M P, Johansen J R. 2014. Seven new species of Oculatella (Pseudanabaenales, Cyanobacteria): taxonomically recognizing cryptic diversification. European Journal of Phycology, 49(4): 450-470.
DOI:10.1080/09670262.2014.976843 |
Papaefthimiou D, Hrouzek P, Mugnai M A, Lukesova A, Turicchia S, Rasmussen U, Ventura S. 2008. Differential patterns of evolution and distribution of the symbiotic behaviour in nostocacean cyanobacteria. International Journal of Systematic and Evolutionary Microbiology, 58(3): 553-564.
DOI:10.1099/ijs.0.65312-0 |
Rajaniemi P, Hrouzek P, Kaštovská K, Willame R, Rantala A, Hoffmann L, Komárek J, Sivonen K. 2005. Phylogenetic and morphological evaluation of the genera Anabaena, Aphanizomenon, Trichormus and Nostoc (Nostocales, Cyanobacteria). International Journal of Systematic and Evolutionary Microbiology, 55(1): 11-26.
DOI:10.1099/ijs.0.63276-0 |
Rambaut A. 2016. Tree figure drawing tool Version 1.4.3. Institute of Evolutionary Biology, United Kingdom, University of Edinburgh. https://vcru.wisc.edu/simonlab/bioinformatics/programs/fgtree/README.txt.
|
Vincent J. 2007. Morphological and molecular characterization of selected desert soil cyanobacteria: three species new to science including Mojavia pulchra gen. et sp. nov. Phycologia, 46: 481-502.
DOI:10.2216/06-92.1 |
Ronquist F, Teslenko M, Van Der Mark P, Ayres D L, Darling A, Höhna S, Larget B, Liu L, Suchard M A, Huelsenbeck J P. 2012. MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61(3): 539-542.
DOI:10.1093/sysbio/sys029 |
Sambrook J, Russell D W. 2001. Molecular cloning: a laboratory manual. 3rd ed. Cold Spring Harbor Laboratory Press, New York. p. 1.84-1.87.
|
Saraf A G, Dawda H G, Singh P. 2019. Validation of the genus Desikacharya gen. nov. (Nostocaceae, Cyanobacteria) and three included species. Notulae Algarum, (107): 1-3.
DOI:10.1099/ijsem.0.001800 |
Saraf A, Dawda H G, Suradkar A, Behere I, Kotulkar M, Shaikh Z M, Kumat A, Batule P, Mishra D, Singh P. 2018. Description of two new species of Aliinostoc and one new species of Desmonostoc from India based on the polyphasic approach and reclassification of Nostoc punensis to Desmonostoc punense comb. nov. FEMS Microbiology Letters, 365(24): fny272.
DOI:10.1093/femsle/fny272 |
Sciuto K, Moro I. 2016. Detection of the new cosmopolitan genus Thermoleptolyngbya (Cyanobacteria, Leptolyngbyaceae) using the 16S rRNA gene and 16S-23S ITS region. Molecular Phylogenetics and Evolution, 105: 15-35.
DOI:10.1016/j.ympev.2016.08.010 |
Silva C S P, Genuário D B, Vaz M G M V, Fiore M F. 2014. Phylogeny of culturable cyanobacteria from Brazilian mangroves. Systematic and Applied Microbiology, 37(2): 100-112.
DOI:10.1016/j.syapm.2013.12.003 |
Stackebrandt E, Ebers J. 2006. Taxonomic parameters revisited: tarnished gold standards. Microbiology Today, 33: 152-155.
|
Stackebrandt E, Goebel B M. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. International Journal of Systematic and Evolutionary Microbiology, 44(4): 846-849.
DOI:10.1099/00207713-44-4-846 |
Taton A, Grubisic S, Brambilla E, De Wit R, Wilmotte A. 2003. Cyanobacterial diversity in natural and artificial microbial mats of Lake Fryxell (McMurdo Dry Valleys, Antarctica): a morphological and molecular approach. Applied and Environmental Microbiology, 69(9): 5157-5169.
DOI:10.1128/AEM.69.9.5157-5169.2003 |
Taton A, Grubisic S, Ertz D, Hodgson D A, Piccardi R, Biondi N, Tredici M R, Mainini M, Losi D, Marinelli F, Wilmotte A. 2006. Polyphasic study of Antarctic cyanobacterial strains. Journal of Phycology, 42(6): 1257-1270.
DOI:10.1111/j.1529-8817.2006.00278.x |
Trifinopoulos J, Nguyen L T, Von Haeseler A, Minh B Q. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Research, 44(W1): W232-W235.
DOI:10.1093/nar/gkw256 |
Turicchia S, Ventura S, Komárková J, Komárek J. 2009. Taxonomic evaluation of cyanobacterial microflora from alkaline marshes of northern Belize. 2. Diversity of oscillatorialean genera. Nova Hedwigia, 89(1-2): 165-200.
DOI:10.1127/0029-5035/2009/0089-0165 |
Vaccarino M A, Johansen J R. 2012. Brasilonema angustatum sp. nov. (Nostocales), a new filamentous cyanobacterial species from the Hawaiian Islands. Journal of Phycology, 48(5): 1178-1186.
DOI:10.1111/j.1529-8817.2012.01203.x |
Watanabe M M, Ichimura T. 1977. Fresh-and salt-water forms of Spirulina platensis in axenic cultures. Bulletin of Japanese Society of Phycology, 25: 371-377.
|
Zapomělová E, Mareš J, Lukešová A, Skácelová O, Řeháková K, Kust A. 2013. Extensive polyphyly of non-gas-vacuolate Anabaena species (Nostocales, Cyanobacteria): a challenge for modern cyanobacterial taxonomy. In: Book of abstracts, 19th Symposium of the International Society for Cyanophyte Research, p. 51, Cleveland, Ohio.
|
 2022, Vol. 40
2022, Vol. 40