Institute of Oceanology, Chinese Academy of Sciences
Article Information
- LI Guoqing, YAN Dingbo, XIA Pinhua, CAO Haipeng, LIN Tao, YI Yin
- Community structure and assembly of denitrifying bacteria in epiphytic biofilms in a freshwater lake ecosystem
- Journal of Oceanology and Limnology, 40(3): 1039-1050
- http://dx.doi.org/10.1007/s00343-021-1084-z
Article History
- Received Mar. 15, 2021
- accepted in principle May. 13, 2021
- accepted for publication May. 31, 2021
2 College of Life Sciences, Guizhou Normal University, Guiyang, 550001, China;
3 State Key Laboratory of Southwest Karst Mountain Biodiversity Conservation of Forestry and Grassland Administration, Guizhou Normal University, Guiyang, 550001, China
Nitrogen is a crucial component of living organisms and a vital nutrient for eutrophication of water bodies (Howarth, 2008). Eutrophication caused by excessive nitrogen and phosphorus from agricultural activities and urbanization has become a severe environmental issue (De-Bashan and Bashan, 2010). Submerged plants have gained a lot of attention recently as they could effectively remove nitrogen and phosphorus from eutrophic water bodies (Søndergaard et al., 2007; Han et al., 2018). Interestingly, stems and leaves surfaces of submerged plant are populated with distinct epiphytic bacteria (Flemming and Wuertz, 2019). A highly diverse nitrogen metabolism mechanism related to nitrogen fixation, nitrification, denitrification, and so on observed in those bacterial communities in stems and leaves surfaces (Levi et al., 2015; Yan et al., 2019). Thus, the self-purification function of the water body can be attributed to the submerged plant-epiphytic bacteria composite system. As per the previous report, submerged macrophytes are a crucial niche for nitrogen cycling bacteria (Coci et al., 2010). Previous studies have underestimated the role of epiphytic microorganisms in the water purification function of submerged plants in artificial or natural systems. However, the underlying mechanism for community composition and structure of epiphytic denitrifying bacteria in aquatic biofilms remains mostly unexplored.
Epiphytic biofilms on the surface of the stems and leaves of submerged plants contain microbes (algae, protists, bacteria, and fungi) and biomolecules (organic, inorganic, and extracellular polymers aggregate), which appears as an adhesion layer of varying thickness (Palmer and White, 1997; Lu et al., 2016). They form a microbial symbiosis with diversified functions and collaborative divisions. The epiphytic biofilm presents an obvious gradient of dissolved oxygen for generating a typical aerobic and hypoxic environment and shaping suitable living conditions for aerobic and anaerobic microorganisms (Sand-Jensen et al., 1985; Ji et al., 2015). Thus, epiphytic biofilm has shown the potential to remove simultaneous nitrogen by aerobic nitrification and anaerobic denitrification. Ribot et al. (2012) validated the crucial role of biofilm in the nitrogen transformation process. A few research studies had demonstrated that a large number of microbes were involved in the nitrogen cycle (Kuypers et al., 2018). Those microbes with related functional genes colonized the epiphytic biofilms on Potamogeton crispus and Wolfia australia (Xie et al., 2015; Yan et al., 2018). Yan et al. (2019) validated outstanding denitrification capabilities of epiphytic bacterial communities using functional prediction analysis. Furthermore, Mu et al. (2020) highlighted the substantial role of epiphytic biofilms in nitrogen removal using isotope labeling.
A myriad of studies have shown that the environmental factors (pH, temperature, light, redox potential, water flow, nutrient availability, and dissolved oxygen concentration) influence the dynamics of epiphytic bacterial communities (Kuehn et al., 2014; Hao et al., 2017). Arnon et al. (2007) reported hydrodynamic force mediated regulation of dissolved oxygen and nitrogen mass transport in the biofilms, which in turn affected the denitrogenation efficiency of the microbes. Recent studies stated that external environmental conditions mediated the regulation of epiphytic bacterial community structure (Bengtsson et al., 2010; He et al., 2014). Epiphytic biofilms showed host-specific microbial communities, which could be due to the complex physical or biochemical characteristics of distinct plant leaves or growth stages (Lachnit et al., 2011). However, as compared to the bacterial community, the community structure and seasonal variation of denitrifying bacteria in epiphytic biofilms remain poorly understood.
Microbial ecology research studies have been centered on the assembly rules of microbial communities and compositional dynamics prediction (Emerson and Gillespie, 2008; Zhou and Ning, 2017). Ecological processes (deterministic and stochastic) affect the assembly of local microbial communities simultaneously (Zhou and Ning, 2017). In aquatic ecosystem, hydrodynamics drives both deterministic and stochastic assembly processes by generating changes in selective pressures and by physically translocating microbes across geographic barriers (Graham et al., 2017). Recent studies stated the dominant role of stochastic processes in microeukaryotic/bacterial community assembly in aquatic ecosystem (Chen et al., 2019; He et al., 2020). Typical physicochemical characteristics of the epiphytic biofilms are liable for its complicated assembly mechanisms. For instance, extracellular polymeric substances serve as a physical barrier creating a "closed microenvironment" to reduce bacteria dispersal ratio (a stochastic process) (Seymour et al., 2017). Concurrently, the complex interactions between bacteria and algae may increase deterministic biotic selection (Foster et al., 2011). However, interaction of stochastic and deterministic processes remains ambiguous. The underlying mechanism for the assembly of the microbial community in copious habitats has been explored (Nemergut et al., 2011). However, none of the studies has explored the ecological mechanisms for the assembly of denitrifying bacterial community in epiphytic biofilms.
Denitrification is a crucial pathway to eliminate nitrogen from eutrophic ecosystems (Ji et al., 2015). Nitrite reductase (nir) catalyzes NO2- reduction to NO, a limiting step in the denitrification process (Henry et al., 2004). The nir gene types, nirS and nirK, are widely distributed in multiple ecosystems. These two genes serve as biomarkers for the identification of denitrifying bacteria (Fan et al., 2016).
The objectives of study were to (i) explore the distribution characteristics of denitrifying bacterial community abundance and structure of the biofilms formed on the submerged macrophytes; (ⅱ) to unveil the effect of environmental factors on dynamics of the denitrifying bacterial community; and (ⅲ) to determine the relative significance of stochastic and deterministic processes in shaping nirS-denitrifying bacterial community assembly. nirS gene was employed as biomarker to investigate the denitrifying bacterial community structure and seasonal changes in epiphytic biofilms using Illumina MiSeq high-throughput sequencing. The null model analysis was performed to assess the roles in stochastic and deterministic processes in the assembly of denitrifying microbial communities, which commonly used to explore the construction mechanisms of a microbial community (Tripathi et al., 2018).
2 MATERIAL AND METHOD 2.1 Sample collectionCaohai Wetland (26°49'N–26°53'N, 104°12'E–104°18'E) located in karst area of Guizhou Province, southwest (SW) China is a national nature reserve since 1990. It is a typical wetland in the Yungui Plateau, 25 km2 in area, 2-m average depth, 2 170 m in altitude, 10.6-℃ average temperature, and 1 000-mm average precipitation (Yan et al., 2019). It is the primary a wintering place for black-necked crane Grus nigricollis that is endemic to China. Aquatic plants in this wetland are Ceratophyllum demersum, Potamogetonaceae, Ottelia acuminata var. acuminate, Myriophyllum verticillatum, and so on. We gathered water and biofilms from nine sites in lakes of Caohai wetland in July and November of 2018 (Fig. 1). For details in site description and physicochemical factors, please see Xia et al. (2020).
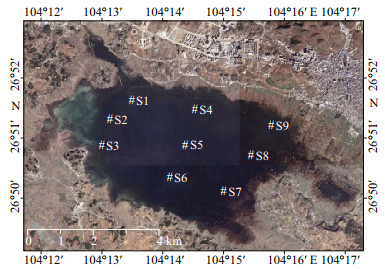
|
| Fig.1 Map of the sample sites (S1–S9) in the Caohai Lake, Guizhou Province, SW China |
The Real-time Quantitative Polymerase Chain Reaction (qPCR) was employed to determine the abundance of nirS gene using the cd3aF (5'-GTSAACGTSAAGGARACSGG-3') and R3cdR (5'-GASTTCGGRTGSGTCTTGA-3') primers (Palmer et al., 2012). F1aCu (5'-ATCATGGTSCTGCCGCG-3') and R3Gu (5'-GCCTCGATCAGRTTGTGGTT-3') were used as the nirK gene primers, as described previously (Fan et al., 2016). PCR products were checked on 1% agarose gel electrophoresis, and the melting curve was constructed to confirm that fluorescent signals were generated only by the PCR products and not from primer-dimers or other artifacts (Yu et al., 2018). The 10-fold serial dilutions of a nirS/nirK-insert containing plasmid were carried out to construct standard curves for qPCR. The gene abundances were calculated from these standard curves (nirS R2=0.999, nirK R2=0.998), and the values were converted to copies per gram of biofilm's fresh weight (BFW), assuming 100% DNA extraction efficiency.
The denitrifying bacteria were identified using primers: cd3aF and R3cdR. The same method was applied in scoring denitrifying bacterial communities from environmental samples (Jung et al., 2011; Yu et al., 2018). Multiple amplification products did not qualify the sequencing quality of the nirK gene, thus the community structure data of the nirK gene were eliminated. All samples were tagged with the forward primer used in the PCR in barcode sequence, which was synthesized using an ABI GeneAmp® 9700 PCR thermocycler (ABI, CA, USA). The 20-μL PCR reaction mixture contained 4 μL of 5X FastPfu Buffer, 0.8 μL of each primer (5 μmol/L), 10 ng of template DNA, 0.4 μL of FastPfu polymerase, BSA 0.2 μL, 2 μL of 2.5 mmol/L dNTPs, and ddH2O up to 20 μL. Amplicons were purified and pooled in equimolar concentration. The paired-end were sequenced on Illumina MiSeq platform (Illumina, San Diego, USA), as per the standard protocols outlined by Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China). Raw sequences were de-multiplexed and quality filtered using QIIME 2. The reads that could not be assembled were discarded. 97% similarity cut-off was used to cluster Operational taxonomic units (OTU) using UPARSE version 7.1 with the novel "greedy" algorithm that simultaneously executes chimera and OTU clustering. The taxonomy of the nirS gene sequence was analyzed by employing the RDP Classifier algorithm (http://rdp.cme.msu.edu/) against the FGR functional gene database with a 70% confidence threshold (Fish et al., 2013). Subsequently, the "vegan" package in R was used to calculate the alpha diversity indexes as per the OTU level. Also, the denitrifying community beta diversity (at OTU level) was as per the Bray-Curtis distances between samples assessed using the "vegan" package in R software (Jiao et al., 2020).
2.3 Data analysisWe divided the growth stages of epiphytic biofilm in July and November into the growth period and decline period, respectively, as detailed in our previous study (Xia et al., 2020). The physicochemical data except for pH were log(x+1) transformed, and the microbial community data were "Hellinger" transformed before analysis (Borcard et al., 2011) for the homoscedasticity and normality. A Wilcoxon test (non-parametric method) was used to compare the seasonal variation of abundance, alpha, and beta diversity of nirS and nirK gene. The Kruskal-Wallis test was employed to determine the significance of nirS and nirK gene abundance, alpha, and beta diversity, and differences among the three submerged macrophytes. The "ggpubr" package in R was utilized for conducting the non-parametric test. The "VennDiagram" package in R compared the differences in the composition of denitrifying bacterial species between growth period and decline period also among three different submerged macrophytes. Non-metric multidimensional scaling analysis (NMDS) and the analysis of similarity (ANOSIM) based on Bray-Curtis distance were employed to visualize the denitrifying bacterial community difference through the "vegan" package in R.
Multiple regressions on distance matrices (MRM), a permutation-based method, unraveled the relationship between variations of bacterial communities in epiphytic biofilms and physicochemical factors of water by 1 000 permutations. Before MRM analysis, using the "ecodist" package, the "bioenv" function in the "vegan" package selected a subset of water's environmental parameters that were best correlated to the community distance (Trumbo et al., 2013). Furthermore, redundancy analysis (RDA) determined the relative importance of physicochemical effect of water, and "ordiR2step" function determined the forward model choice based solely on adjusted R2 and P-value by 1 000 permutations in the "vegan" package.
Relative influences of ecological processes were estimated using an operational framework described by Stegen et al. (2015). The between-community nearest taxon index (βNTI) and Bray-Curtis-based Raup-Crick (RCbray) were calculated through null model-based phylogenetic and taxonomic beta diversity metrics. The null model expectation used 999 randomizations. The classified ecological processes influencing community assembly were categorized as homogeneous selection (βNTI < -2), variable selection (βNTI > 2), homogenizing dispersal (-2 < βNTI < 2 and RCbray < -0.95), dispersal limitation (-2 < βNTI < 2 and RCBray > 0.95), and undominated (-2 < βNTI < 2 and -0.95 < RCBray < 0.95, a condition for which no single process dominates community assembly).
3 RESULT 3.1 Abundance of nirS and nirK geneThe nirS and nirK gene abundance was summarized in Fig. 2. The number of nirS gene copy ranged from 5.54×106 to 3.89×107 copies per gram of BFW, and 2.92×106 to 1.13×107 copies per gram of BFW during the growth period and decline period of the plant, respectively (P < 0.05). The gene abundance of nirK ranged from 6.71×106 to 9.76×108 copies per gram of BFW and 3.44×106 to 1.93×107 copies per gram of BFW during the growth period and decline period of the plant, respectively, which was higher than nirS gene abundance (P < 0.05). Substantial differences (P < 0.05) in the abundance of epiphytic denitrification genes in different host plants were observed, as depicted in Fig. 2. The numbers of nirS and nirK gene copies in the epiphytic biofilm on the P. lucens were the highest during the growth period (2.79×107±1.96×107 and 7.53×106±8.41×106 copies per gram of BFW, respectively) and decline period (7.76×108±5.78×108 and 2.91×107±6.67×107 copies per gram of BFW, respectively). The nirS and nirK gene copy number in the epiphytic biofilms of the Potamogeton lucens and M. verticillatum decreased from growth period to decline period, while epiphytic biofilms on the C. demersum were increased.

|
| Fig.2 Bubble diagram showing the abundance (copies/g BFW) of the denitrification genes in epiphytic biofilm a. nirS; b. nirK. |
Rare bacterial species were not considered in this analysis. Deficient reads were removed except for those who had ≥ 5 sequence reads in at least three samples or that 10% of the total samples contained these reads. 382 OTUs of denitrifying bacteria containing the nirS gene were identified from 641 656 quality sequences at 97% similarity level for the 32 samples (samples JS1_1, NS6_1, and NS4_2 were eliminated due to poor sequencing quality). Supplementary Table S1 summarizes the Good's coverage and alpha diversity of the denitrifying bacterial community in the epiphytic biofilm at the OTU level of 97% similarity. The Good's coverage ranged from 0.997 to 0.999 in the individual samples, and the integrated index of all the 32 samples was 0.998, indicating that the majority of the denitrifying bacterial taxa was identified from metacommunity. The abundance-based coverage estimator (ACE), Shannon-Weiner index, phylogenetic diversity (PD), and Pielou's evenness did not change significantly between the growth period and decline period of macrophyte. ACE and Shannon index were slightly higher in winter than in summer. In addition, in each period, no sizeable differences were observed in the alpha diversity of the denitrifying bacterial community from the three host plants.
Most of the nirS-denitrifying bacteria belonged to phylum Proteobacteria (56.55%±22.15%), and others were unclassified bacteria (41.71%±22.17%) (Fig. 3), amounting up to 91.39%–99.98% of the total reads from different samples. Proteobacteria growth showed no significant difference between growth period (July) and decline (November) period (54.50% vs. 58.54%), while the Gammaproteobacteria growth was faster during growth period than the decline period (P < 0.005). The relative abundance of Proteobacteria in epiphytic biofilm on M. verticillatum was the high during both the growth period and decline period (Fig. 3).
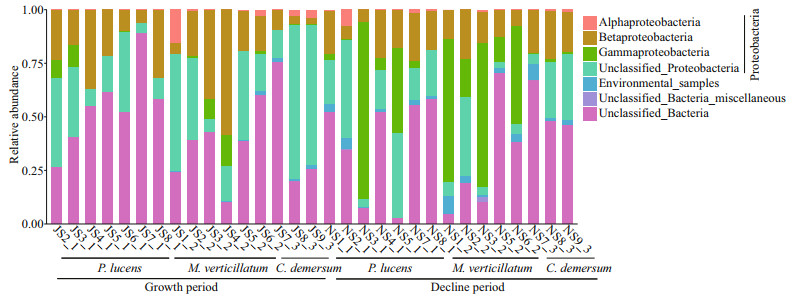
|
| Fig.3 The relative abundance of different denitrifying bacteria in biofilms formed on Potamogeton lucens, Myriophyllum verticillatum, and Ceratophyllum demersum |
Variances in composition of the nirS-denitrifying bacterial communities between growth period and decline period and among host plants were investigated using the Venn diagram analysis at OTU level (Fig. 4). We found that 76.18% of the total 291 OTUs were shared between the two periods in the case of nirS-denitrifying bacteria. Meanwhile, epiphytic biofilms formed during the plant growth period was twice of the decline period. Three submerged macrophytes had 132 common OTUs (37.5%) of the epiphytic denitrifying bacterial community during the growth period and 136 OTUs (42.37%) during the decline period. The denitrifying bacteria in epiphytic biofilms of P. lucens and M. verticillatum had the highest OTU sharing rate of 72.76% during the growth period and 64.84% during the decline period. P. lucens showed the highest unique OTUs during growth period and decline period.
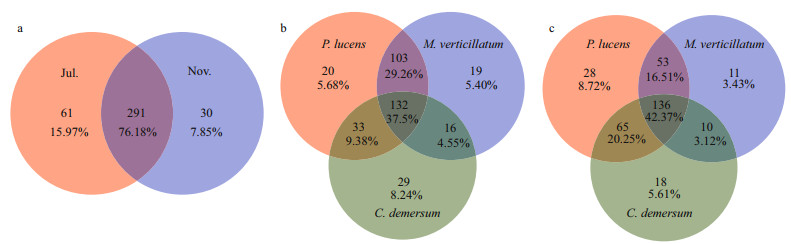
|
| Fig.4 The Venn diagram of the denitrifying bacterial community (OTU level) showing species in common a. in July and November; b & c. common species in three plants in July (b) and in November (c), respectively. |
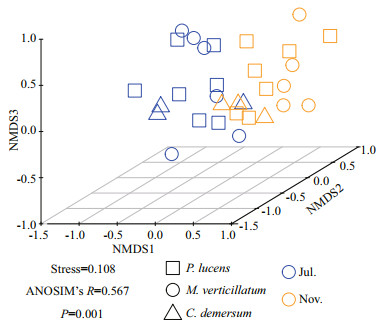
The beta diversity of denitrifying bacterial communities was summarized in Fig. 5. Beta diversity (P < 0.000 1) of denitrifying bacteria was significantly higher in epiphytic biofilms during the growth period than in the decline period. Conversely, beta diversity was significantly higher in epiphytic biofilms during the decline period than the growth period in P. lucens (P < 0.001), whereas M. verticillatum showed the opposite trend. To unveil the differences in nirS-denitrifying bacterial community structure between growth period and decline period, an NMDS analysis was performed based on the Bray-Curtis distance as per the OTU distribution (Fig. 6). The denitrifying bacterial community structure during the growth period was remarkably different from the decline period (Stress=0.108). ANOSIM analysis further proved the variance in the denitrifying bacterial community structure between growth period and decline period (R=0.567, P=0.001). As shown in this analysis, denitrifying bacterial community structure was different in three submerged macrophytes during growth period and decline period (R=0.259, P=0.029 for growth period, and R=0.218, P=0.049 for decline period) (Fig. 6).
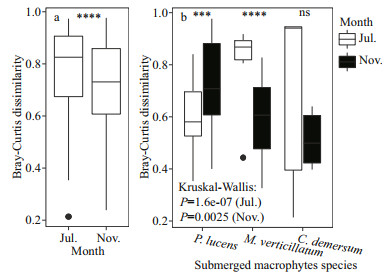
|
| Fig.5 The box-diversity plots of biofilm denitrification functional communities in different plants in different months a. difference in diversity between different seasons; b. difference in diversity among different host plants. ***: P < 0.001; ****: P < 0.000 1; ns: not significant; black dots represent outliers. |
|
| Fig.6 The NMDS similarities between all samples |
The Mantel test presented significant correlation between physiochemical factors, such as water temperature (WT), disolved oxygen (DO), pH, total phosphorus (TP), NH4+, and the nirS-denitrifying bacterial communities of the submerged macrophytes during the growth period (Mantel's R=0.435, P=0.007; Supplementary Table S2). However, during the decline period, WT, pH, total nitrogen (TN), and chlorophyll a (Chl a) were significantly correlated to the nirS-denitrifying bacterial communities (Mantel's R=0.299, P=0.015). Thus, denitrifying bacterial communities showed significant seasonal variation in correlation with WT, DO, and NO3- (Mantel's R=0.477, P=0.001). Multiple regression analysis indicated that using physicochemical factors of water column could not discern the diversity in denitrifying bacterial community from epiphytic biofilms (MRM's R2 was 0.14, 0.103, and 0.238 for growth period, decline period, and both, respectively). Alternatively, the RDA results demonstrated that variation in a few denitrifying bacterial communities could be explained through water parameters (R2 was 0.182, 0.281, and 0.36 for the growth period, decline period, and both, respectively; Supplementary Table S3).
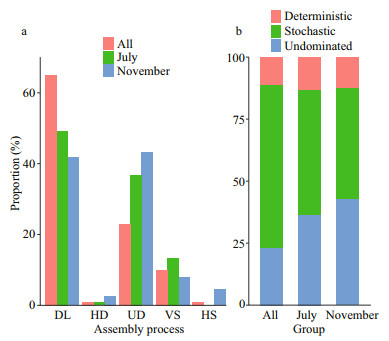
3.5 Phylogeny-based null model analysisAs indicated by the relative contribution of ecological process, physicochemical factors of water column played a minor role in the assembly of denitrifying bacterial community in epiphytic biofilm (Fig. 7), while dispersal limitation process played a major role in shaping the denitrifying community structure in both growth period (49.22%) and decline period (41.78%). In addition, seasonal-change-related effect was crucial on the denitrifying bacterial community in epiphytic biofilms (64.52%). Secondly, the undominated process played an essential role in the assembly of denitrifying bacterial community during the growth period, decline period, and seasonal changes. Furthermore, the subdivision of ecological processes indicated that the stochastic process had a higher contribution than the deterministic process in growth period and decline period, including the seasonal changes. Notably, the undominated process played a minor role in community assembly in all seasons.
|
| Fig.7 Proportions of denitrifying bacteria assembled by diffusion-limited (DL), homogeneous diffusion (HD), uncontrolled (UD), factor selection (VS), and homogeneous selection (HS) according to the null model analysis (a); the proportion of denitrification communities assembled from deterministic, stochastic, and undominated processes (b) |
In the current study, we observed a higher nirS and nirK gene abundance during the growth period (July) than the decline period (November), which was in line with the previous studies (Lindemann et al., 2016; Huang et al., 2018). Multiple reports accentuated the fact that environmental factors affect the abundance and activity of genes involved in the biogeochemical nitrogen cycle (Sunagawa et al., 2015). As stated in our previous study on the abundance of epiphytic bacteria, temperature affects substantially the microbial abundance (Xia et al., 2020), and so the gene abundance. In this study, the epiphytic denitrification genes were isolated from the microbiome present on leaf surfaces of three submerged macrophyte species. We observed that the abundance of nirK gene was higher than that of the nirS gene in summer and winter. Previous studies have shown that the abundance of nirK gene was higher in multiple environmental samples, such as sediment (Saarenheimo et al., 2015), epiphytic biofilms (Vila-Costa et al., 2014), soil (Dandie et al., 2011), and so on than that of nirS. It may be due to the oxygen-rich microenvironment of biofilms, which might have hampered the growth of oxygen-sensitive nirS-type bacteria and promoted the growth of aerobic nirK-type bacteria (Knapp et al., 2009). We also observed that the abundance of the nirK gene was higher in the growth period than the decline period. In addition, higher variation of nirK gene abundance between different seasons than that of nirS gene can be seen in Fig. 2. It was suggested that the nirK-type denitrifying bacteria were more sensitive to environmental variations than that of the nirS-type denitrifying bacteria (Dandie et al., 2011; Yuan et al., 2012).
A myriad of studies demonstrated denitrification activity in the submerged epiphytic biofilms of macrophytes in freshwater ecosystem (Fan et al., 2016; Yan et al., 2019). Interestingly, the denitrification gene abundances in biofilms was higher than in other denitrification spots, such as sediments and water columns. The most probable number (MPN) method was widely employed to calculate the density of denitrifying bacteria in submerged macrophytes (Körner, 1999; Eriksson, 2001). Wuhle River (Berlin, Germany) that serves as a dumping site for the effluent of Falkenberg sewage treatment plant, showed the denitrifying bacterial density of 9.4×104 to 7×104 MPN/mL in water and 3.3×106 to 2.5×106 MPN/cm3 in sediment. The average number of denitrifying bacteria was higher (8.7×107±1×107 MPN per gram of plant dry weight) in the epiphytic biofilms of 6 submerged macrophytes including Ceratophyllum demersum, Potamogeton natans L., Potamogeton crispus L., Sagittaria sagittifolia L., Sparganium emersum rehmann L., and Potamogeton pectinatus L. (Körner, 1999). In the Donghu (East) Lake, a eutrophic lake in Wuhan, Hubei Province, the abundance of nirS gene ranged from 1.47×104 to 1.67×105 copies per gram of dry sediment, and that of nirK gene ranged from 2.02×107 to 9.71×107 copies per gram of dry sediment (Hou et al., 2013). However, the abundance of nirS and nirK gene obtained in our analysis was significantly higher than that of the Donghu Lake. Besides, Fan et al. (2016) reported a higher abundance of nirS and nirK genes in epiphytic bacterial communities of plants Potamogeton malaianus and C. demersum than the water samples from Taihu Lake. Mu et al. (2020) identified the critical contribution of epiphytic biofilms of Myriophyllum spicatum in nitrogen removal by isotope labeling. Thus, biofilms on leaves of submerged macrophytes in aquatic ecosystem have outstanding denitrification potential. These are a novel approach for controlling nitrogen levels in aquatic ecosystem in the foreseeable future.
In this study, most of the denitrification microbes in the biofilms belonged to phylum Proteobacteria (56.55%±22.15%) while the rest of the bacteria were unclassified (41.71%±22.17%) (Fig. 3). Previous phylogenetic analysis showed that the aerobic denitrifying bacteria mainly belonged to Alpha-, Beta-, and Gamma-Proteobacteria (Ji et al., 2015). Occurrence of diverse aerobic denitrifying bacteria and alternating anaerobic-oxygenic environments in biofilms on plants (Sand-Jensen et al., 1985; Ji et al., 2015) created a conducive environment for denitrification. In addition, epiphytic biofilms can uptake a sea of nitrogen nutrients in the surrounding water (Levi et al., 2015), and the nitrogen could be further utilized for the development of epiphytic biofilms and microbial growth involved in the nitrogen cycle (Levi et al., 2015; Yan et al., 2018). Moreover, secretion from the plant leaf surface provide a carbon substrate for microbes in the nitrogen cycle (Florez et al., 2017), and epiphytic algae respiration increases the pH of the epiphytic biofilms. The microenvironmental factors, such as physical barrier (diffusion limitation) of extracellular polymer in the biofilms, enhanced the denitrification function (Yan et al., 2019). The specific microbial community composition and microenvironment of epiphytic biofilms promote the nitrogen removal from the water.
Denitrifying microbial community in epiphytic biofilms showed the highest species commonality during the growth period and decline period. However, differences in the structure of the denitrifying microbial community between growth period and decline period were apparent (R=0.5, P=0.001). Concurrently, according to the outcomes of the functional gene quantification (Fig. 2), the denitrifying bacterial density in epiphytic biofilms during the growth period was significantly higher than that of the decline period. Thus, we believe that difference in the denitrifying microbial community structure of epiphytic biofilm was due primarily to depleted microbial abundance. First, higher biomass in July, i.e., the plant growth period, was resulted from water temperature, which stimulated the growth and reproduction of microbes (Bengtsson et al., 2010). Secondly, strong photosynthesis of plants during growth period and sufficient oxygen and carbohydrates accelerated the growth and reproduction of aerobic microbes (Yan et al., 2019). However, in November (the plant decline period), as the ambient temperature dropped, the growth and metabolism of microbes were also depleted. Also, the decreased photosynthetic activity of the host plant hampered the growth of the primary dominant species and introduced or promoted the growth of organic-matter-decomposing bacteria (Doughari et al., 2011; Mancuso et al., 2016; Xia et al., 2020). These bacteria also went through the selection pressure of the complex organic molecules released by the plant during the decline period (Seymour et al., 2017; Zhang et al., 2018). At the same time, biofilms gradually matured from July to November, drawing more nutrients from water. Yan et al. (2018) demonstrated that a higher nutrient concentration led to the inhibition of denitrifying microorganisms and the reduction of the biomass in the biofilms. Finally, the abundance and activity of the denitrifying bacteria in the epiphytic biofilm was depleted.
Temperature was the key factor in limiting the seasonal dynamic characteristics of microbial communities as per previous studies (Casartelli and Ferragut, 2015). In this study, MRM, Mantel test, and RDA analysis indicated that temperature was the primary factor on seasonal dynamics of denitrifying microbes in epiphytic biofilms. Our study of the effect of aquatic environmental factors on the dynamics of biofilm's denitrifying microbial communities indicated that multiple factors affected the assembly process of denitrifying microbial communities. It implies the involvement of biotic and abiotic factors on the epiphytic denitrifying bacterial community. These factors, such as epiphytic biofilm, algal secretion, and respiration, increased the pH in the ambience of epiphytic biofilms, promoted the buildup of epiphytic microbial community and the heterotrophic microbial activity, and affected the ecological function of the microbial community (Kuehn et al., 2014; Song et al., 2015). Also, the low effect of aquatic environmental factors implies that the stochastic process may be the vital driving force to the construction of denitrifying microbial communities. Results of the null model analysis (Fig. 7) indicated that the dynamic characteristics of the denitrifying microbial community were affected by stochastic dispersal limitation (64.52%) and undominated (23.20%) processes. The underlying driving mechanism for microbial community assembly in aquatic ecosystems had been controversial. Previous research reports revealed that the deterministic process forms bacterioplankton communities (Sunagawa et al., 2015; Wang et al., 2019), while stochastic process regulates microbial community structure (Evans et al., 2017; Zhou and Ning, 2017).
In this study, the biofilms created a nearly closed microenvironment, and the physical barrier made of extracellular polymer limited the microbial dispersion (Liu et al., 2016; Wu, 2016). Many microbes, such as Alphaproteobacteria and Betaproteobacteria, do not have motor flagella and thus show low motor ability (Roger et al., 2017). This suggests that the construction of denitrifying bacteria community was significantly influenced by the morphological (type, size, and shape) and habitat (openness, geographical distance) of the bacterial community. It was in line with the size-dispersal hypothesis (a stochastic process) (Liu et al., 2020) and distance-decay patterns (a stochastic process) (Astorga et al., 2012). Overall, the community assembly of nirS-type bacteria was influenced by both stochastic and deterministic processes, but the stochastic processes such as dispersal limitation emerged as the dominant mechanism as per our analysis.
5 CONCLUSIONAbundant denitrification genes was found in the biofilm formed on the surface of leaves of submerged plants with seasonal changes. Meanwhile, abundance of nirK gene was higher than that of nirS gene in biofilm. In addition, nirS gene showed more environmental stability than nirK gene did. Proteobacteria was the dominant phylum in the nirS-type denitrifying bacterial community in the epiphytic biofilms and showed distinct seasonal differences. The nirS-type denitrifying community dynamics could not be well explained by environmental factors assessed in this study. Combining the null model analysis results, we suggested that stochastic and deterministic processes affected the community assembly of nirS-type bacteria. And the stochastic process, such as dispersal limitation, emerged as the principal mechanism in this study. However, the process of temporal changes in the abundance, composition, structure and assembly process of denitrifying bacteria from the growth period (July) to decline period (November) of the epiphytic biofilms remains unresolved. Therefore, research on the denitrification function and community dynamic mechanism of the epiphytic biofilms demands further investigations.
6 DATA AVAILABILITY STATEMENTThe data that support the finding of this study are available from the corresponding author.
7 ACKNOWLEDGMENTThe authors thank the editor and reviewers in helping to improve the manuscript. We also thank Xu SONG, Yu YANG, Xiangchen TANG, Tianyou WANG, Xin DU, and Mengmeng FAN from Guizhou Province Key Laboratory for Information System of Mountainous Areas and Protection of Ecological Environment for their generous help in sampling and laboratory works.
Electronic supplementary materialSupplementary materials (Supplementary Tables S1–S3) are available in the online version of this article at https://doi.org/10.1007/s00343-021-1084-z.
Arnon S, Peterson C G, Gray K A, Packman A I. 2007. Influence of flow conditions and system geometry on nitrate use by benthic biofilms: implications for nutrient mitigation. Environmental Science & Technology, 41(23): 8142-8148.
DOI:10.1021/es0710048 |
Astorga A, Oksanen J, Luoto M, Soininen J, Virtanen R, Muotka T. 2012. Distance decay of similarity in freshwater communities: do macro- and microorganisms follow the same rules?. Global Ecology and Biogeography, 21(3): 365-375.
DOI:10.1111/j.1466-8238.2011.00681.x |
Bengtsson M M, Sjøtun K, Øvreas L. 2010. Seasonal dynamics of bacterial biofilms on the kelp Laminaria hyperborea. Aquatic Microbial Ecology, 60(1): 71-83.
DOI:10.3354/ame01409 |
Borcard D, Gillet F, Legendre P. 2011. Numerical Ecology with R. Springer, New York, NY. 435p.
|
Casartelli M R, Ferragut C. 2015. Influence of seasonality and rooted aquatic macrophyte on periphytic algal community on artificial substratum in a shallow tropical reservoir. International Review of Hydrobiology, 100(5-6): 158-168.
DOI:10.1002/iroh.201401773 |
Chen W D, Ren K X, Isabwe A, Chen H H, Liu M, Yang J. 2019. Stochastic processes shape microeukaryotic community assembly in a subtropical river across wet and dry seasons. Microbiome, 7(1): 138.
DOI:10.1186/s40168-019-0749-8 |
Coci M, Nicol G W, Pilloni G N, Schmid M, Kamst-Van Agterveld M P, Bodelier P L E, Laanbroek H J. 2010. Quantitative assessment of ammonia-oxidizing bacterial communities in the epiphyton of submerged macrophytes in shallow lakes. Applied and Environmental Microbiology, 76(6): 1813-1821.
DOI:10.1128/AEM.01917-09 |
Dandie C E, Wertz S, Leclair C L, Goyer C, Burton D L, Patten C L, Zebarth B J, Trevors J T. 2011. Abundance, diversity and functional gene expression of denitrifier communities in adjacent riparian and agricultural zones. FEMS Microbiology Ecology, 77(1): 69-82.
DOI:10.1111/j.1574-6941.2011.01084.x |
De-Bashan L E, Bashan Y. 2010. Immobilized microalgae for removing pollutants: review of practical aspects. Bioresource Technology, 101(6): 1611-1627.
DOI:10.1016/j.biortech.2009.09.043 |
Doughari H J, Ndakidemi P A, Human I S, Benade S. 2011. The ecology, biology and pathogenesis of Acinetobacter spp.: an overview. Microbes and Environments, 26(2): 101-112.
DOI:10.1264/jsme2.ME10179 |
Emerson B C, Gillespie R G. 2008. Phylogenetic analysis of community assembly and structure over space and time. Trends in Ecology & Evolution, 23(11): 619-630.
DOI:10.1016/j.tree.2008.07.005 |
Eriksson P G. 2001. Interaction effects of flow velocity and oxygen metabolism on nitrification and denitrification in biofilms on submersed macrophytes. Biogeochemistry, 55(1): 29-44.
DOI:10.1023/A:1010679306361 |
Evans S, Martiny J B H, Allison S D. 2017. Effects of dispersal and selection on stochastic assembly in microbial communities. The ISME Journal, 11(1): 176-185.
DOI:10.1038/ismej.2016.96 |
Fan Z, Han R M, Ma J, Wang G X. 2016. Submerged macrophytes shape the abundance and diversity of bacterial denitrifiers in bacterioplankton and epiphyton in the shallow fresh Lake Taihu, China. Environmental Science and Pollution Research, 23(14): 14102-14114.
DOI:10.1007/s11356-016-6390-1 |
Fish J A, Chai B L, Wang Q, Sun Y N, Brown C T, Tiedje J M, Cole J R. 2013. FunGene: the functional gene pipeline and repository. Frontiers in Microbiology, 4: 291.
DOI:10.3389/fmicb.2013.00291 |
Flemming H C, Wuertz S. 2019. Bacteria and archaea on Earth and their abundance in biofilms. Nature Reviews Microbiology, 17(4): 247-260.
DOI:10.1038/s41579-019-0158-9 |
Florez J Z, Camus C, Hengst M B, Buschmann A H. 2017. A functional perspective analysis of macroalgae and epiphytic bacterial community interaction. Frontiers in Microbiology, 8: 2561.
DOI:10.3389/fmicb.2017.02561 |
Foster R A, Kuypers M M M, Vagner T, Paerl R W, Musat N, Zehr J P. 2011. Nitrogen fixation and transfer in open ocean diatom-cyanobacterial symbioses. The ISME Journal, 5(9): 1484-1493.
DOI:10.1038/ismej.2011.26 |
Graham E B, Crump A R, Resch C T, Fansler S, Arntzen E, Kennedy D W, Fredrickson J K, Stegen J C. 2017. Deterministic influences exceed dispersal effects on hydrologically-connected microbiomes. Environmental Microbiology, 19(4): 1552-1567.
DOI:10.1111/1462-2920.13720 |
Han B, Zhang S H, Zhang L S, Liu K H, Yan L Y, Wang P F, Wang C, Pang S. 2018. Characterization of microbes and denitrifiers attached to two species of floating plants in the wetlands of Lake Taihu. PLoS One, 13(11): e0207443.
DOI:10.1371/journal.pone.0207443 |
Hao B B, Wu H P, Cao Y, Xing W, Jeppesen E, Li W. 2017. Comparison of periphyton communities on natural and artificial macrophytes with contrasting morphological structures. Freshwater Biology, 62(10): 1783-1793.
DOI:10.1111/fwb.12991 |
He D, Ren L J, Wu Q L. 2014. Contrasting diversity of epibiotic bacteria and surrounding bacterioplankton of a common submerged macrophyte, Potamogeton crispus, in freshwater lakes. FEMS Microbiology Ecology, 90(3): 551-562.
DOI:10.1111/1574-6941.12414 |
He R J, Zeng J, Zhao D Y, Huang R, Yu Z B, Wu Q L. 2020. Contrasting patterns in diversity and community assembly of Phragmites australis root-associated bacterial communities from different seasons. Applied and Environmental Microbiology, 86(14): e00379-20.
DOI:10.1128/AEM.00379-20 |
Henry S, Baudoin E, López-Gutiérrez J C, Martin-Laurent F, Brauman A, Philippot L. 2004. Quantification of denitrifying bacteria in soils by nirK gene targeted realtime PCR. Journal of Microbiological Methods, 59(3): 327-335.
DOI:10.1016/j.mimet.2004.07.002 |
Hou J, Cao X Y, Song C L, Zhou Y Y. 2013. Predominance of ammonia-oxidizing archaea and nirK-gene-bearing denitrifiers among ammonia-oxidizing and denitrifying populations in sediments of a large urban eutrophic lake (Lake Donghu). Canadian Journal of Microbiology, 59(7): 456-464.
DOI:10.1139/cjm-2013-0083 |
Howarth R W. 2008. Coastal nitrogen pollution: A review of sources and trends globally and regionally. Harmful Algae, 8(1): 14-20.
DOI:10.1016/j.hal.2008.08.015 |
Huang S, Chen C, Jaffé P R. 2018. Seasonal distribution of nitrifiers and denitrifiers in urban river sediments affected by agricultural activities. Science of The Total Environment, 642: 1282-1291.
DOI:10.1016/j.scitotenv.2018.06.116 |
Ji B, Yang K, Zhu L, Jiang Y, Wang H Y, Zhou J, Zhang H N. 2015. Aerobic denitrification: a review of important advances of the last 30 years. Biotechnology and Bioprocess Engineering, 20(4): 643-651.
DOI:10.1007/s12257-015-0009-0 |
Jiao S, Yang Y F, Xu Y Q, Zhang J, Lu Y H. 2020. Balance between community assembly processes mediates species coexistence in agricultural soil microbiomes across eastern China. The ISME Journal, 14(1): 202-216.
DOI:10.1038/s41396-019-0522-9 |
Jung J, Yeom J, Kim J, Han J, Lim H S, Park H, Hyun S, Park W. 2011. Change in gene abundance in the nitrogen biogeochemical cycle with temperature and nitrogen addition in Antarctic soils. Research in Microbiology, 162(10): 1018-1026.
DOI:10.1016/j.resmic.2011.07.007 |
Knapp C W, Dodds W K, Wilson K C, O'Brien J M, Graham D W. 2009. Spatial heterogeneity of denitrification genes in a highly homogenous urban stream. Environmental Science & Technology, 43(12): 4273-4279.
DOI:10.1021/es9001407 |
Körner S. 1999. Nitrifying and denitrifying bacteria in epiphytic communities of submerged macrophytes in a treated sewage channel. Acta Hydrochimica et Hydrobiologica, 27(1): 27-31.
DOI:10.1002/(sici)1521-401x(199901)27:1<27::aid-aheh27>3.0.co;2-1 |
Kuehn K A, Francoeur S N, Findlay R H, Neely R K. 2014. Priming in the microbial landscape: periphytic algal stimulation of litter-associated microbial decomposers. Ecology, 95(3): 749-762.
DOI:10.1890/13-0430.1 |
Kuypers M M M, Marchant H K, Kartal B. 2018. The microbial nitrogen-cycling network. Nature Reviews Microbiology, 16(5): 263-276.
DOI:10.1038/nrmicro.2018.9 |
Lachnit T, Meske D, Wahl M, Harder T, Schmitz R. 2011. Epibacterial community patterns on marine macroalgae are host-specific but temporally variable. Environmental Microbiology, 13(3): 655-665.
DOI:10.1111/j.1462-2920.2010.02371.x |
Levi P S, Riis T, Aknøe A B, Peipoch M, Maetzke K, Bruus C, Baattrup-Pedersen A. 2015. Macrophyte complexity controls nutrient uptake in lowland streams. Ecosystems, 18(5): 914-931.
DOI:10.1007/s10021-015-9872-y |
Lindemann S, Zarnoch C B, Castignetti D, Hoellein T J. 2016. Effect of Eastern Oysters (Crassostrea virginica) and Seasonality on Nitrite Reductase Gene Abundance (nirS, nirK, nrfA) in an Urban Estuary. Estuaries and Coasts, 39(1): 218-232.
DOI:10.1007/s12237-015-9989-4 |
Liu J Z, Wang F W, Liu W, Tang C L, Wu C X, Wu Y H. 2016. Nutrient removal by up-scaling a hybrid floating treatment bed (HFTB) using plant and periphyton: from laboratory tank to polluted river. Bioresource Technology, 207: 142-149.
DOI:10.1016/j.biortech.2016.02.011 |
Liu K S, Liu Y Q, Hu A Y, Wang F, Chen Y Y, Gu Z Q, Anslan S, Hou J Z. 2020. Different community assembly mechanisms underlie similar biogeography of bacteria and microeukaryotes in Tibetan lakes. FEMS Microbiology Ecology, 96(6): fiaa071.
DOI:10.1093/femsec/fiaa071 |
Lu H Y, Wan J J, Li J Y, Shao H B, Wu Y H. 2016. Periphytic biofilm: a buffer for phosphorus precipitation and release between sediments and water. Chemosphere, 144: 2058-2064.
DOI:10.1016/j.chemosphere.2015.10.129 |
Mancuso F P, D'hondt S, Willems A, Airoldi L, De Clerck O. 2016. Diversity and temporal dynamics of the epiphytic bacterial communities associated with the canopy-forming seaweed Cystoseira compressa (Esper) Gerloff and Nizamuddin. Frontiers in Microbiology, 7: 476.
DOI:10.3389/fmicb.2016.00476 |
Mu X Y, Lv X Y, Liu W, Qiu C H, Ma Y, Zhang S H, Jeppesen E. 2020. Biofilms attached to Myriophyllum spicatum play a dominant role in nitrogen removal in constructed wetland mesocosms with submersed macrophytes: evidence from 15N tracking, nitrogen budgets and metagenomics analyses. Environmental Pollution, 266: 115203.
DOI:10.1016/j.envpol.2020.115203 |
Nemergut D R, Costello E K, Hamady M, Lozupone C, Jiang L, Schmidt S K, Fierer N, Townsend A R, Cleveland C C, Stanish L, Knight R. 2011. Global patterns in the biogeography of bacterial taxa. Environmental Microbiology, 13(1): 135-144.
DOI:10.1111/j.1462-2920.2010.02315.x |
Palmer Jr R J, White D C. 1997. Developmental biology of biofilms: implications for treatment and control. Trends in Microbiology, 5(11): 435-440.
DOI:10.1016/S0966-842X(97)01142-6 |
Palmer K, Biasi C, Horn M A. 2012. Contrasting denitrifier communities relate to contrasting N2O emission patterns from acidic peat soils in arctic tundra. The ISME Journal, 6(5): 1058-1077.
DOI:10.1038/ismej.2011.172 |
Ribot M, Martí E, Von Schiller D, Sabater F, Daims H, Battin T J. 2012. Nitrogen processing and the role of epilithic biofilms downstream of a wastewater treatment plant. Freshwater Science, 31(4): 1057-1069.
DOI:10.1899/11-161.1 |
Roger A J, Muñoz-Gómez S A, Kamikawa R. 2017. The origin and diversification of mitochondria. Current Biology, 27(21): R1177-R1192.
DOI:10.1016/j.cub.2017.09.015 |
Saarenheimo J, Rissanen A J, Arvola L, Nykänen H, Lehmann M F, Tiirola M. 2015. Genetic and environmental controls on nitrous oxide accumulation in lakes. PLoS One, 10(3): e0121201.
DOI:10.1371/journal.pone.0121201 |
Sand-Jensen K, Revsbech N P, Jørgensen B B. 1985. Microprofiles of oxygen in epiphyte communities on submerged macrophytes. Marine Biology, 89(1): 55-62.
DOI:10.1007/BF00392877 |
Seymour J R, Amin S A, Raina J B, Stocker R. 2017. Zooming in on the phycosphere: the ecological interface for phytoplankton-bacteria relationships. Nature Microbiology, 2(7): 17065.
DOI:10.1038/nmicrobiol.2017.65 |
Søndergaard M, Jeppesen E, Lauridsen T L, Skov C, Van Nes E H, Roijackers R, Lammens E, Portielje R. 2007. Lake restoration: successes, failures and long-term effects. Journal of Applied Ecology, 44(6): 1095-1105.
DOI:10.1111/j.1365-2664.2007.01363.x |
Song Y Z, Wang J Q, Gao Y X, Xie X J. 2015. The physiological responses of Vallisneria natans to epiphytic algae with the increase of N and P concentrations in water bodies. Environmental Science and Pollution Research, 22(11): 8480-8487.
DOI:10.1007/s11356-014-3998-x |
Stegen J C, Lin X J, Fredrickson J K, Konopka A E. 2015. Estimating and mapping ecological processes influencing microbial community assembly. Frontiers in Microbiology, 6: 370.
DOI:10.3389/fmicb.2015.00370 |
Sunagawa S, Coelho L P, Chaffron S, Kultima J R, Labadie K, Salazar G, Djahanschiri B, Zeller G, Mende D R, Alberti A, Cornejo-Castillo F M, Costea P I, Cruaud C, D'ovidio F, Engelen S, Ferrera I, Gasol J M, Guidi L, Hildebrand F, Kokoszka F, Lepoivre C, Lima-Mendez G, Poulain J, Poulos B T, Royo-Llonch M, Sarmento H, Vieira-Silva S, Dimier C, Picheral M, Searson S, Kandels-Lewis S, Coordinators T O, Bowler C, Vargas C D, Gorsky G, Grimsley N, Hingamp P, Iudicone D, Jaillon O, Not F, Ogata H, Pesant S, Speich S, Stemmann L, Sullivan M B, Weissenbach J, Wincker P, Karsenti E, Raes J, Acinas S G, Bork P. 2015. Structure and function of the global ocean microbiome. Science, 348(6237): 1261359.
DOI:10.1126/science.1261359 |
Tripathi B M, Stegen J C, Kim M, Dong K, Adams J M, Lee Y K. 2018. Soil pH mediates the balance between stochastic and deterministic assembly of bacteria. The ISME Journal, 12(4): 1072-1083.
DOI:10.1038/s41396-018-0082-4 |
Trumbo D R, Spear S F, Baumsteiger J, Storfer A. 2013. Rangewide landscape genetics of an endemic Pacific northwestern salamander. Molecular Ecology, 22(5): 1250-1266.
DOI:10.1111/mec.12168 |
Vila-Costa M, Bartrons M, Catalan J, Casamayor E O. 2014. Nitrogen-cycling genes in epilithic biofilms of oligotrophic high-altitude lakes (central Pyrenees, Spain). Microbial Ecology, 68(1): 60-69.
DOI:10.1007/s00248-014-0417-2 |
Wang K, Hu H J, Yan H Z, Hou D D, Wang Y T, Dong P S, Zhang D M. 2019. Archaeal biogeography and interactions with microbial community across complex subtropical coastal waters. Molecular Ecology, 28(12): 3101-3118.
DOI:10.1111/mec.15105 |
Wu Y H. 2016. Periphyton: Functions and Application in Environmental Remediation. Elsevier, Netherlands. 402p.
|
Xia P H, Yan D B, Sun R G, Song X, Lin T, Yi Y. 2020. Community composition and correlations between bacteria and algae within epiphytic biofilms on submerged macrophytes in a plateau lake, southwest China. Science of the Total Environment, 727: 138398.
DOI:10.1016/j.scitotenv.2020.138398 |
Xie W Y, Su J Q, Zhu Y G. 2015. Phyllosphere bacterial community of floating macrophytes in paddy soil environments as revealed by Illumina high-throughput sequencing. Applied and Environmental Microbiology, 81(2): 522-532.
DOI:10.1128/AEM.03191-14 |
Yan D B, Xia P H, Song X, Lin T, Cao H P. 2019. Community structure and functional diversity of epiphytic bacteria and planktonic bacteria on submerged macrophytes in Caohai Lake, southwest of China. Annals of Microbiology, 69(9): 933-944.
DOI:10.1007/s13213-019-01485-4 |
Yan L Y, Zhang S H, Lin D, Guo C, Yan L L, Wang S P, He Z L. 2018. Nitrogen loading affects microbes, nitrifiers and denitrifiers attached to submerged macrophyte in constructed wetlands. Science of the Total Environment, 622-623: 121-126.
DOI:10.1016/j.scitotenv.2017.11.234 |
Yu Z H, Liu J J, Li Y S, Jin J, Liu X B, Wang G H. 2018. Impact of land use, fertilization and seasonal variation on the abundance and diversity of nirS-type denitrifying bacterial communities in a Mollisol in Northeast China. European Journal of Soil Biology, 85: 4-11.
DOI:10.1016/j.ejsobi.2017.12.001 |
Yuan Q, Liu P F, Lu Y H. 2012. Differential responses of nirK- and nirS-carrying bacteria to denitrifying conditions in the anoxic rice field soil. Environmental Microbiology Reports, 4(1): 113-122.
DOI:10.1111/j.1758-2229.2011.00311.x |
Zhang L S, Zhang S H, Lv X Y, Qiu Z, Zhang Z Q, Yan L Y. 2018. Dissolved organic matter release in overlying water and bacterial community shifts in biofilm during the decomposition of Myriophyllum verticillatum. Science of the Total Environment, 633: 929-937.
DOI:10.1016/j.scitotenv.2018.03.275 |
Zhou J Z, Ning D L. 2017. Stochastic community assembly: does it matter in microbial ecology?. Microbiology and Molecular Biology Reviews, 81(4): e00002-17.
DOI:10.1128/MMBR.00002-17 |
 2022, Vol. 40
2022, Vol. 40


