Institute of Oceanology, Chinese Academy of Sciences
Article Information
- WU Sha, WANG Qing, WANG Xu, GUO Ruixue, ZHANG Tongwei, PAN Yongxin, LI Feng, LI Ying
- MamZ protein plays an essential role in magnetosome maturation process of Magnetospirillum gryphiswaldense MSR-1
- Journal of Oceanology and Limnology, 39(6): 2082-2096
- http://dx.doi.org/10.1007/s00343-021-0321-9
Article History
- Received Aug. 27, 2020
- accepted in principle Sep. 29, 2020
- accepted for publication Dec. 28, 2020
2 State Key Laboratories for Agro-biotechnology, China Agricultural University, Beijing 100193, China;
3 Paleomagnetism and Geochronology Laboratory, Key Laboratory of Earth and Planetary Physics, Institute of Geology and Geophysics, Chinese Academy of Sciences, France-China Joint Laboratory for Evolution and Development of Magnetotactic Multicelluar Organisms, Beijing 100029, China
Magnetotactic bacteria (MTB) are characterized by the ability to synthesize nano-scale magnetic particles ("magnetosomes") that consist of a lipid bilayer and Fe3O4 or Fe3S4 (Frankel et al., 1979; Balk et al., 1980; Gorby et al., 1988; Heywood et al., 1990). Biosynthesis of magnetosomes is strictly controlled by a series of genes located mainly on a "magnetosome island" (MAI) (Schüler, 2004). In Magnetospirillum gryphiswaldense strain MSR-1, MAI consists of feoAB1, mamAB, mamFDC, mamXY, and mms6 operons (Wang et al., 2014). According to Murat et al. (2010), biomineralization of magnetosomes in M. magnetotacticum strain AMB-1 involves five steps: (i) invagination of magnetosome vesicle from inner membrane, (ii) sorting of magnetosome proteins to magnetosome membrane, (iii) alignment of magnetosomes in a chain along a filamentous cytoskeletal structure, (iv) iron uptake and initiation of crystal formation, (v) crystal maturation. Each of these steps involves certain key proteins, and the steps are not entirely distinct from each other. The functions of MAI genes have been the focus of increasing study during the past decade. In 2014, Schüler's group characterized the MAI genes in Rhodospirillum rubrum and their roles in magnetosome biosynthesis (Kolinko et al., 2014). To better understand the molecular mechanism of magnetosome biomineralization, functions of specific MAI genes in other species and strains of MTB remain to be elucidated.
The present study was focused on four genes located in the mamXY operon of MSR-1: mamY, mamX, mamZ, and ftsZ-like (Wang et al., 2014).MamY contains the Bin/amphiphysin/Rvs (BAR) domain, which displays direct binding activity to liposomes composed of phospholipids. In sedimentation assay, MamY was co-precipitated with the liposomes. Magnetosome vesicles in mamY deletion mutant were larger than in wild-type (WT) strain (Tanaka et al., 2010). These findings suggest that MamY functions to constrict the magnetosome membrane during vesicle formation. MamX contains two CXXCH heme-binding motifs typical of c-type cytochromes (Raschdorf et al., 2013; Yang et al., 2013). FtsZ-like displayed both ATPase and GTPase activities, resulting in GTP-dependent polymerization into tubulin filament bundles in vitro (Ding et al., 2010). ftsZ-like and mamX deletion mutants formed small, superparamagnetic particles (Ding et al., 2010; Yang et al., 2013) that are similar to those of entire mamXY operon deletion mutant (Lohße et al., 2011).
mamX and mamZ have a 32-bp region of sequence overlap in MSR-1 based on genome sequence and analysis in 2014 (Wang et al., 2014). The analysis of mamX revealed a close relationship with mamZ, and complementary functions among the proteins encoded by mamXY operon (Yang et al., 2013). Schüler's group proposed that MamZ, MamX, and MamH functionally interact in the magnetosome membrane to form an iron oxidoreductase / transport complex for magnetite biomineralization (Raschdorf et al., 2013). Nudelman and Zarivach (2014) presented bioinformatics analysis and structure predictions of important MAI genes that were consistent with our previous findings (Ding et al., 2010; Yang et al., 2013). However, visual data of this sort must be confirmed by various physiological and biochemical analyses.
In this study, we successfully constructed a mamZ-deletion mutant strain (ΔmamZ) and four complemented strains with differing mamZ fragment lengths. The effects of mamZ deletion on cell phenotypic and physiological characteristics in MSR-1, and on transcription levels of other genes were evaluated. Comparative analysis of the characteristics of these strains helped elucidate the function of mamZ, and the relationships among key MAI genes.
2 MATERIAL AND METHOD 2.1 Bioinformatics analysisTwo templates were used for 3D structure prediction of the major facilitator superfamily (MFS) and the ferric-reduction superfamily (FRS) domains of MamZ: glycerol-3-phosphate transporter (GlpT), belonging to the MFS transport family from Escherichia coli (PDB ID: 1PW4), and partialcytochrome b of the cytochrome bc1 complex (complex III) from Saccharomyces cerevisiae (PDB ID: 3CX5). Homologue-modeling structures of the two domains were generated using the SWISS-MODEL web server following target sequencing and uploading of corresponding template PDB format files. Generated PDB files were loaded into PyMOL molecular visualization system to produce visual protein models.
2.2 Bacterial strains and growth conditionsBacterial strains and plasmids used in this study are listed in Table 1. MSR-1 WT and mutant strains were cultured in sodium lactate medium (SLM) at 30 ℃ with 100 r/min shaking (Rong et al., 2008). SLM was autoclaved before sterile ferric citrate added. E. coli cells were cultured in Luria broth (LB) at 37 ℃ with 200 r/min shaking. Antibiotics used in culture media were as follows: for MSR-1 strains, gentamicin (Gm), tetracycline (Tc), and nalidixic acid (Nx) all at 5 μg/mL; for E. coli, ampicillin (Amp) at 100 μg/mL, Gm at 20 μg/mL, kanamycin (Km) at 50 μg/mL, and Tc at 12.5 μg/mL.
A mamZ deletion mutant of MSR-1 (termed ΔmamZ) was constructed by conjugation and subsequent homologous recombination. The primer sequences used are listed in Supplementary Table S1. Construction of ΔmamZ is illustrated in Supplementary Fig.S1, and method described as previous (Yang et al., 2013). PCR primers for each fragment length to construction of complemented strains are listed in Supplementary Table S1. The respective truncated mamZ genes were cloned into pRK415 and then introduced into ΔmamZ by biparental conjugation. The four correct complemented strains (confirmed by rigorous examination) were termed CmamZ (full length mamZ; 1 947 bp; 648 aa), CpmamZ (partial fragment; 281–648 aa), CMFS (N-terminal; MFS superfamily; 1–449 aa), and CredFe (C-terminal; ferric-reduction superfamily; 471–629aa), respectively.
2.4 Cell growth curve and magnetic response (Cmag)Cells were cultured in SLM at 30 ℃ with 100 r/min shaking (Wang et al., 2015a). Cell growth (OD565) and Cmag for each strain were measured at various sampling times as described previously (Wang et al., 2015a).
2.5 Transmission electron microscopy (TEM)Cells were collected at 24 h, placed on copper or carbon grids, washed thrice with double-distilled H2O, and observed by TEM (model JEM-1230, JEOL; Tokyo, Japan) or high-resolution TEM (HR-TEM) (model JEM-2100, JEOL). Numbers and diameters of magnetosomes were analyzed statistically using ImageJ (National Institutes of Health; Bethesda, MD, USA), a Java-based image-processing program. Any two groups were compared with the analysis of independent sample T test by IBM SPSS Statistics.
2.6 Rock magnetic measurementsFor rock magnetic measurements (Yang et al., 2013), room-temperature hysteresis loops and first-order reversal curves (FORC) of cells were measured by an alternating gradient force magnetometer (model MicroMag 3900, Princeton Measurements Corp.; Princeton, NJ, USA; sensitivity 5.0×10-10 Am2) (Wang et al., 2015b).
2.7 Iron content and iron uptake of experimental strainsStrains were grown in SLM supplemented with 20-or 60- mol/L ferric citrate at 30 ℃ for 24 h, and harvested by centrifugation (8 000×g, 5 min). The precipitate was digested by nitric acid as described previously (Rong et al., 2008; Wang et al., 2013), and total intracellular iron content was assayed by inductively coupled plasma optical emission spectrometry (ICP-OES) (model Optima 5300DV, Perkin Elmer; Waltham, MA, USA).
Strains were cultured in 100-mL SLM with 60-μmol/L ferric citrate and centrifuged at various sampling times. The supernatant was used for detection of iron concentration remaining in the medium by the ferrozine standard curve method (Wang et al., 2013). Ferric citrate was prepared in various concentrations, reduced to ferrous ions by hydroxylamine hydrochloride, reacted with ferrozine as a chromogenic reagent, and detected at OD562. A standard curve was generated based on values obtained. Residual iron concentration (mol/L Fe2+) in supernatant was measured and calculated at each sampling time using the standard curve.
2.8 RNA extraction and RT-qPCRCells were cultured in 100-mL SLM and harvested by centrifugation (12 000×g, 1 min, 4 ℃). Total RNA was purified using TRIzol reagent (Tiangen Biotech; Beijing, China) according to the manufacturer's instructions. Remaining genomic DNA in RNA was digested with RNase-free DNase I (TaKaRa, Japan). Quality and quantity of the extraction were assessed by A 260 nm/A280 nm ratio.
cDNA was obtained as described in our previous study (Wang et al., 2013). RT-qPCR was performed using a LightCycler 480 RT-PCR system and LightCycler 480 SYBR Green I Master Kit (Roche; Mannheim, Germany). The primers used for amplification of MAI genes are listed in Supplementary Table S2. The rpoC gene encoding RNA polymerase subunit β' was chosen as internal control and reference. Transcription levels were analyzed by the 2-ΔΔCt method (Wang et al., 2013).
3 RESULT 3.1 Structural and functional prediction of mamZ geneOrientation and composition of mamZ, based on bioinformatics analysis, are summarized in Fig. 1. mamZ contains 1 947 bp, encoding 648 aa residues.The genome number of mamZ in MSR-1 is MGMSRv2_2323 (Fig. 1a). MamZ has 18 transmembrane helices (Fig. 1b) containing two conserved domains. The 12 N-terminal helices belong to MFS (449 aa; Fig. 1c), and have 64% identity with MamH in MTB. MFS, member of the large class of secondary active transporters presenting in cells of all kingdoms, is predicted to utilize the electrochemical potential of one substance to drive the transport of another substance (Madej et al., 2013; Madej and Kaback, 2013; Yan, 2013). The 6 C-terminal helices belong to FRS (159 aa; Fig. 1c), and have high identity with YedZ in E. coli (Richter et al., 2007; Raschdorf et al., 2013; Nudelman and Zarivach, 2014). In E. coli, YedZ (a membrane-intrinsic cytochrome b with six putative transmembrane helices) and YedY (a soluble catalytic subunit with a twin arginine leader sequence) form a YedYZ complex that is a sulfite oxidase homologue (von Rozycki et al., 2004; Brokx et al., 2005; Drew et al., 2005). MamZ appears to be responsible for iron transport using the N-terminal MFS region and ferric-reduction-related redox reaction of iron using the C-terminal FRS region. Sites for interaction with other DNAs, RNAs, and proteins are scattered throughout the gene sequence, but concentrated mainly in the C-terminus, particularly the 281–648 aa region (partial MamZ; Fig. 1c).
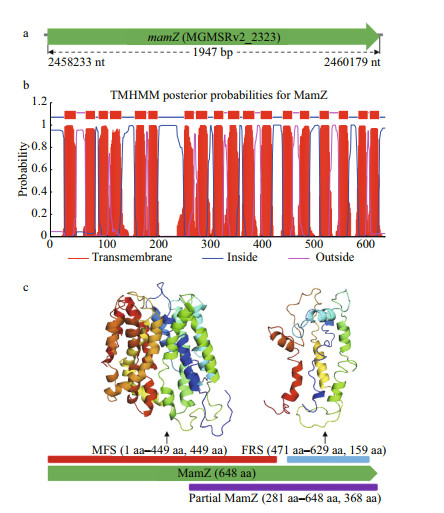
|
| Fig.1 Protein structure analysis of MamZ a. length of mamZ gene (gene number: MGMSRv2_2323), and its location in MSR-1 genome; b. transmembrane prediction of MamZ by TMHMM method. X-axis: aa number of MamZ protein from N-terminal to C-terminal; Y-axis: probability of transmembrane for each aa. Red blocks: possible transmembrane regions; blue and pink lines: peptide fragments inside and outside membrane; c. schematic diagram of MamZ and its specific domain/region. Green arrow: MamZ protein; orientation: sequence from N-terminal to C-terminal; red and blue rectangles: two predicted domains of MFS and FRS; black arrows: protein structures of two domains generated through homologue modeling; purple rectangle: MamZ region covering FRS domain and partial MFS domain. |
Taking into account the multiple transmembrane domains of MamZ, we considered that complementation of the full-length gene would be difficult and problematic. Hence, we divided the complementary gene fragment into encoding full-length MamZ (1–648 aa), N-terminal MFS (1– 449 aa), C-terminal FRS (471–629 aa), and part of the extension of the C-terminal regions (partial MamZ, 281–648 aa). The associated experimental design is summarized in Fig. 1c.
3.2 mamZ deletion has no effect on cell growth, but results in loss of magnetic responseCareful examination and sequencing analysis confirmed that the deletion mutant (ΔmamZ) strain showed no change relative to mamZ at either the left or right side, thus ruling out a polarity effect. WT, ΔmamZ, and four complemented strains (integrity of MAI genes was ensured in each strain) were cultured in SLM supplemented with 20- or 60-μmol/L ferric citrate. Compared to WT, except that ΔmamZ and CredFe lagging ~6 h, the growth rates of the other strains (CmamZ, CpmamZ, and CMFS) did not differ notably (Fig. 2a & c). Moreover, the maximal OD565 of various strains distributed in 1.5–1.7 range. Cmag values for the strains are shown in Fig. 2b & d. For WT under two iron concentrations, maximal Cmag values were observed at 20 h (0.788 under 20-μmol/L Fe3+; Fig. 2b) and 14 h (1.10 under 60-μmol/L Fe3+; Fig. 2d). Cmag value was zero for ΔmamZ throughout the incubation period, regardless of iron concentration.For full-length complemented strain CmamZ, maximal Camg values were 0.06 under 20-μmol/L and 0.10 under 60-μmol/L ferric citrate conditions that generally 10% of WT (Fig. 2b & d). Under both conditions, Cmag values of CmamZ lagged ~2 h behind that of WT (Fig. 2b & d). However, Cmag values of the other complemented strains (CpmamZ, CMFS, and CredFe) were the same as that of ΔmamZ. Thus, segmental mamZ was insufficient to ΔmamZ to generate enough Cmag value. RT-qPCR analysis showed that mamZ transcription level was ~10-fold lower in CmamZ than in WT (Supplementary Fig S2). This result is consistent with Cmag values as compared to that of WT. The expression of full-length and segmental mamZ in the four complemented strains was reduced obviously, which might be another reason of the low Cmag values.
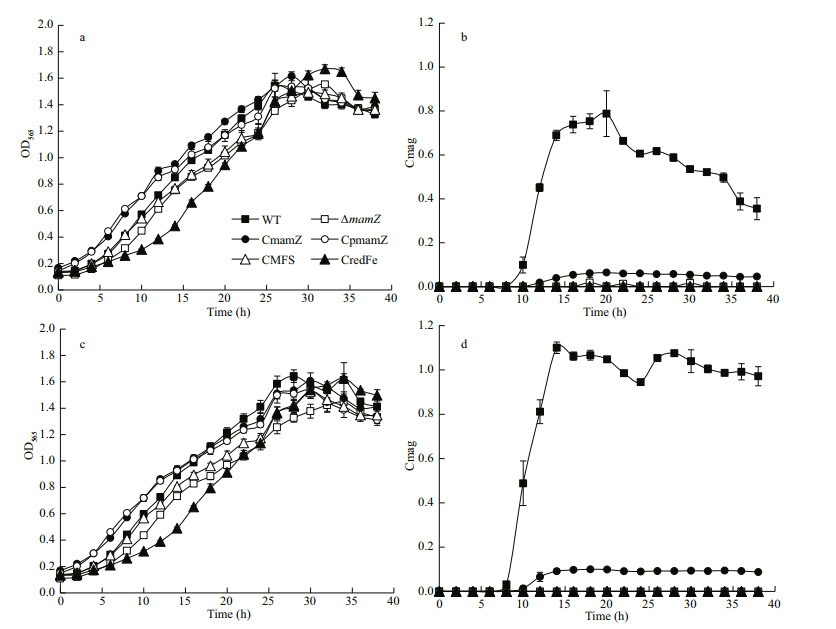
|
| Fig.2 Comparison of cell growth and magnetic response (Cmag) in six strains cultured with 20-μmol/L (a, b) and 60-μmol/L (c, d) ferric citrate Growth rates were not notably different among the six strains under 20-μmol/L (a) or 60-μmol/L ferric citrate (c) condition. Cmag value (b, d) of ΔmamZ was consistently zero under the two conditions. Cmag value of CmamZ was ~10% of WT value under both conditions. Cmag values of the other three complemented strains were zero under both conditions. All experiments were performed in triplicate. |
Magnetosomes in the six strains (WT, ΔmamZ, CmamZ, CpmamZ, CMFS, CredFe) were observed by HR-TEM. WT cells contained regular cubo-octahedral magnetosomes with mean diameter 28.22±0.45 nm and typical magnetite crystal lattice (Table 2; Fig. 3a–c). In ΔmamZ, compared to WT, magnetosomes had lower mean number (11.89±1.068), smaller mean diameter (17.58±0.304 nm) (Table 2; Fig. 3d & e), and were more irregular with poor crystal lattice (Fig. 3f). Magnetosomes in CmamZ (Fig. 3g–i), CpmamZ (Fig. 3j–l), CMFS (Fig. 3m–o), and CredFe (Fig. 3p–r) were irregular and smaller than in WT, whereas mean magnetosome numbers in the four complemented strains were higher than in WT. The complemented strains evidently lacked the ability to synthesize mature magnetosomes, and instead formed many abnormal, smaller crystals (Table 2). It implied that cells lost the ability to synthesize normal magnetosomes due to mamZ expression deleted or disrupted.
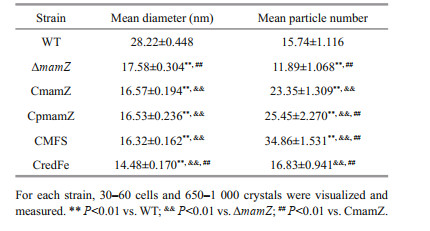
|
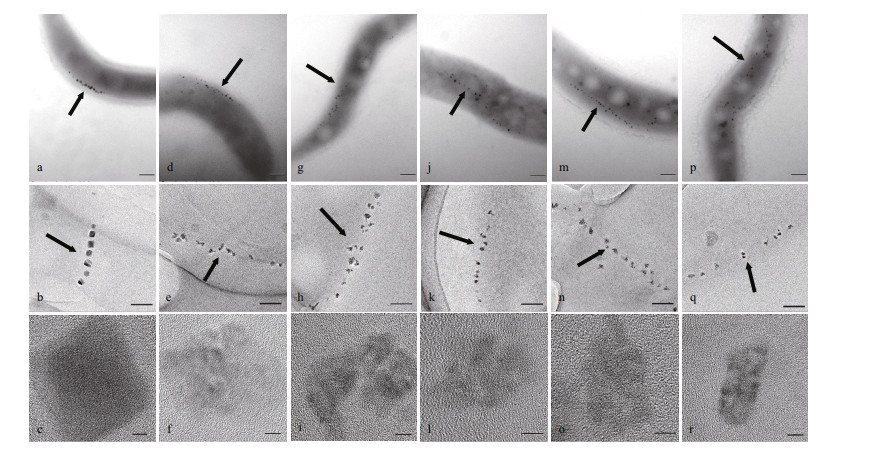
|
| Fig.3 TEM and HR-TEM observations of six strains Panels show WT (a, b, c), ΔmamZ (d, e, f), and complemented strains (CmamZ: g, h, i; CpmamZ: j, k, l; CMFS: m, n, o; CredFe: p, q, r). Top row (a, d, g, j, m, p): cell phenotype. Arrows: magnetosome chains. Scale bars: 200 nm. Middle row (b, e, h, k, n, q): magnetosome chain organization. Scale bars: 100 nm.Bottom row (c, f, i, l, o, r): crystal lattice structure. Scale bars: 5 nm. mamZ deletion resulted in small, irregular, and poor-lattice particles. |
The six strains were cultured with shaking for 24 h, and cells were harvested and collected by centrifugation. Magnetic properties of these minerals were analyzed using room-temperature hysteresis loops and FORC as described in Section 2. WT displays a typical Stoner-Wohlfarth type loop with Hc, Hcr, Hcr/Hc, and Mrs/Ms values of 14.18 mT, 19.39 mT, 1.37, and 0.40, respectively (Fig. 4a1). The FORC diagram for WT (Fig. 4a2) have closed inner contours centered around ~20 mT along the horizontal axis and with narrow distribution along the vertical axis, which further indicates the dominance of stable-single-domain-like particles. However, the hysteresis loop and FORC diagrams became noisy when mamZ was deleted (Fig. 4b1 & b2), indicating the extremely weak magnetism of cells. Apparent immature particles were still detected in some ΔmamZ cells by HR-TEM, but were in the form of poor-lattice. Compared with WT, the coercivity values of CmamZ, CpmamZ, CMFS, and CredFe decreased to 4.39, 7.59, 4.08, and 10.67 mT, respectively (Fig. 4c1, d1, f1), and FORC diagrams have closed contours for single domain particles centered below 20 mT and some outer contours for superparamagnetic particles perpendicular to the vertical axis close to the origin of FORC diagrams (Fig. 4c2, d2, e2, f2). These results indicated that the four kinds of complemented strains synthesized magnetic particles, but the magnetism of complemented strains was significantly weaker than the magnetism of WT. That is mamZ expression, whether full-length or segmental, could repair ΔmamZ part capability of magnetosome biosynthesis. However, the magnetism of magnetic particles biosynthesized by complemented strains fails to reach that of WT.
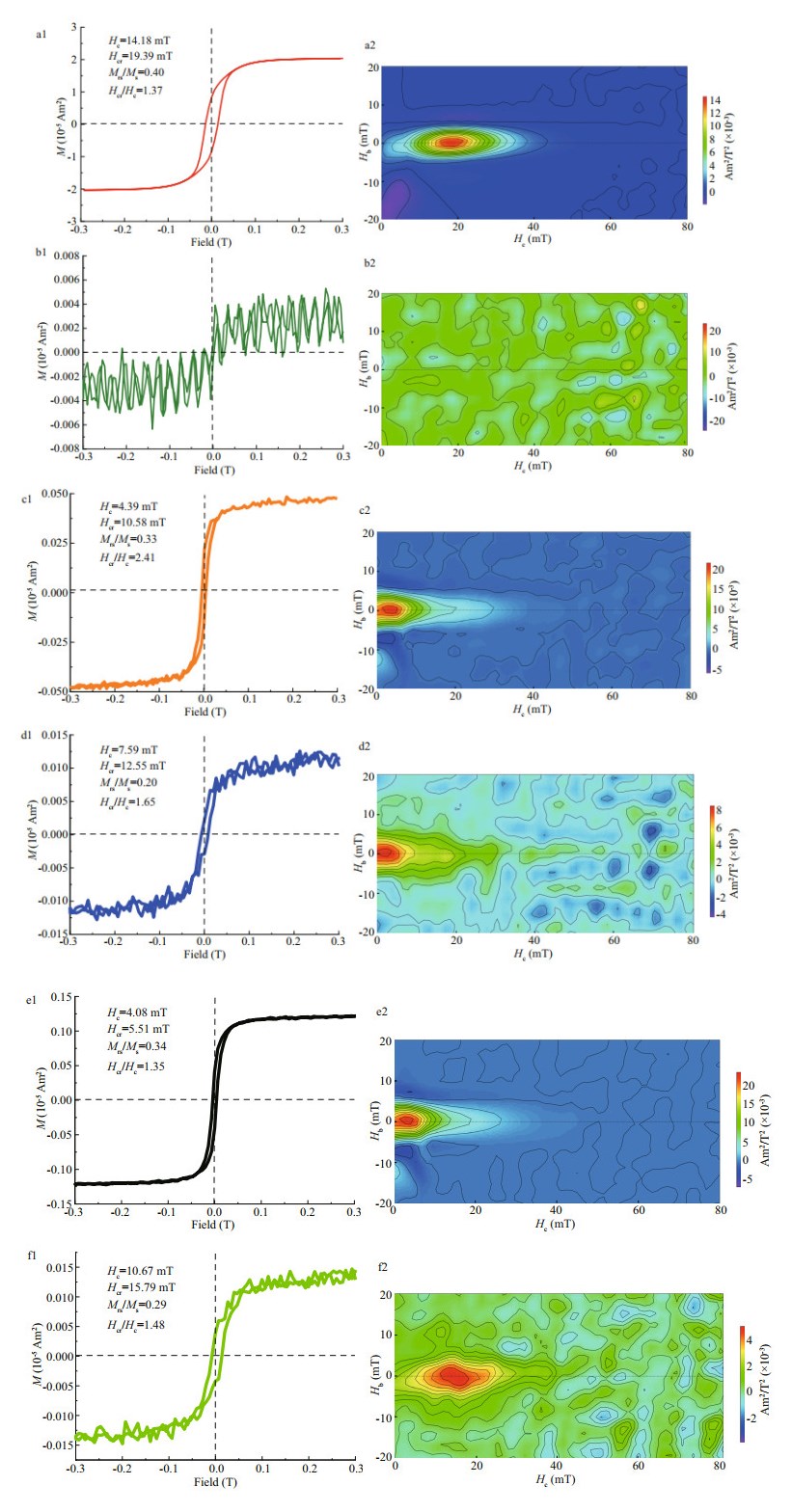
|
| Fig.4 Room-temperature hysteresis loop and FORC for six strains WT cells (a1, a2) contain single-domain (SD) magnetosomes. ΔmamZ cells (b1, b2) show very weak magnetism of smaller and immature magnetosomes. CmamZ cells (c1, c2), CpmamZ (d1, d2), CMFS (e1, e2), and CredFe (f1, f2) contain mainly immature particles. |
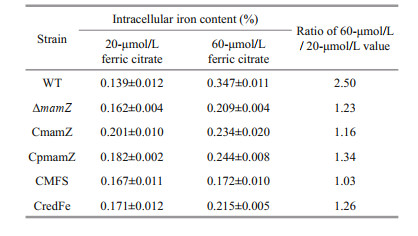
Previous studies demonstrated that 20- mol/L iron concentration is sufficient for normal magnetosome biosynthesis in MSR-1 (Rong et al., 2008), but that cells could tolerate higher concentrations (Zhang et al., 2013). To further characterize ΔmamZ and the four complemented strains, we measured iron uptake ability of cells cultured in media containing 20- and 60-μmol/L ferric citrate. Under 20-μmol/L ferric citrate condition, intracellular iron content was slightly higher in ΔmamZ and the complemented strains than in WT, but otherwise did not differ notably among the various strains (Table 3). For WT, iron content was 2.5-fold higher under 60-μmol/L compared to 20-μmol/L ferric citrate condition, indicating that iron uptake was stimulated by increasing iron concentration in medium. The other strains showed iron content enhancement in the 1.0-to 1.3-fold range (Table 3). In regard to iron uptake, the strains were next compared under 60- mol/L ferric citrate condition, including sampling at regular intervals of cell growth (OD565) and testing residual iron ion content in medium. Iron content in medium decreased gradually over time for all strains; however, residual iron content in medium during the culture process was highest for ΔmamZ (Supplementary Fig. S3). Thus, mamZ deletion resulted in reduced iron uptake when cells were cultured under high iron concentration. Among the complemented strains, residual iron content in medium was lower for CmamZ and CMFS than for CpmamZ and CredFe at each sampling time (Supplementary Fig.S3). This finding is presumably due to the fact CmamZ and CMFS contain the integrated MFS domain of MamZ. Transformation of MFS domain into ΔmamZ enhanced iron uptake under high-iron condition, consistent with predictions of MamZ structure.
|
To evaluate effects of mamZ deletion on other genes in the mamXY operon, we measured relative transcription levels of mamY, mamX, and ftsZ-like in the six strains at different growth stages, e.g., the six strains were cultured for 12 h (logarithmic phase of cell growth; OD565 ~0.45) and for 18 h (early stage of stationary phase; OD565 ~0.80). Relative transcription level of mamY was slightly higher in ΔmamZ, CmamZ, CMFS, and CredFe than in WT at 12 h and 18 h, and at 18 h was 1- to 2-fold higher in CpmamZ than in the other strains (Fig. 5a). mamX level at 12 h was slightly higher in ΔmamZ, CmamZ, and CredFe than in WT (Fig. 5b). Compared to 12 h, mamX level at 18 h was enhanced significantly in WT, but to a lesser degree in the other strains (Fig. 5b). ftsZ-like level was upregulated moderately at 12 h in ΔmamZ and the complemented strains, and significantly (1- to 5-fold) at 18 h, particularly in CmamZ, CpmamZ, and CMFS (Fig. 5c). Hence, the relative transcription levels of mamY, mamX, and ftsZ-like in ΔmamZ or thecomplemented strains were upregulated with varying degrees. In the model of MamXY proteins during magnetosome formation, MamZ was expressed at last stage and formed complexes with MamX and FtsZ-like (Wang et al., 2019). However, there had no interaction between MamX and MamZ, and they were separately recruited by FtsZ-like at different stages, respectively. It is a pity that only one interaction site was detected in MamZ with FtsZ-like. This might be the reason that ftsZ-like expressed most significant, especially at 18 h in CmamZ, CMFS, and CpmamZ, the next was mamX, and the last was mamY. Two of the mamXY operon genes (mamX and ftsZ-like) were clearly upregulated during the stationary phase of cell growth, which proved the corresponds to the gradual maturation stage of magnetosomes once again.

|
| Fig.5 Relative transcription levels of mamY, mamX, and ftsZ-like in WT, ΔmamZ, and the four complemented strains incubated for 12 h and 18 h Transcription levels of mamX and (particularly) ftsZ-like were upregulated at 18 h for all strains, indicating that the encoded proteins play essential roles in the magnetosome maturation process. |
There is sequence overlap (32 bp) between mamX and mamZ (Wang et al., 2014), and deletion of either gene led to formation of similar poor-lattice magnetosomes (Yang et al., 2013). In addition to ΔmamZ and its complemented, we therefore performed additional experiments on ΔmamX and its complemented strain (CmamX), simultaneously.
The MAI consists of feoAB1, mamAB, mamFDC, mamXY, and mms6 operons. We randomly selectedimportant genes located in operons other than mamXY, and determined relative transcription levels of these genes in ΔmamZ and its complemented strains. The seven selected genes, and primers used, are listed in Supplementary Table S2. Transcription level of mms6 was much higher than that of the other genes in ΔmamZ and the four complemented strains at both 12 h (Fig. 6a) and 18 h (Fig. 6b). In particular, mms6 transcription level was 10- to 50-fold higher than those of the other genes in ΔmamZ at 12 h, and > 50-fold higher in the complemented strains at 18 h. Levels of mamB and mamF were slightly enhanced in CredFe at 12 h, and those of mamF and mamE were two-fold higher in CmamZ, CpmamZ, and CMFS at 18 h. Similar, the results were obtained for CmamX (Fig. 6c & d). Levels of certain genes were higher in complemented strains; e.g., mamF, mamN, mamB increased ~2-fold. mms6 level was increased 15-fold in CmamX at 12 h (Fig. 6c), and 12-fold at 18 h (Fig. 6d). The other genes did not show notable changes relative to WT at 18 h (Fig. 6d).
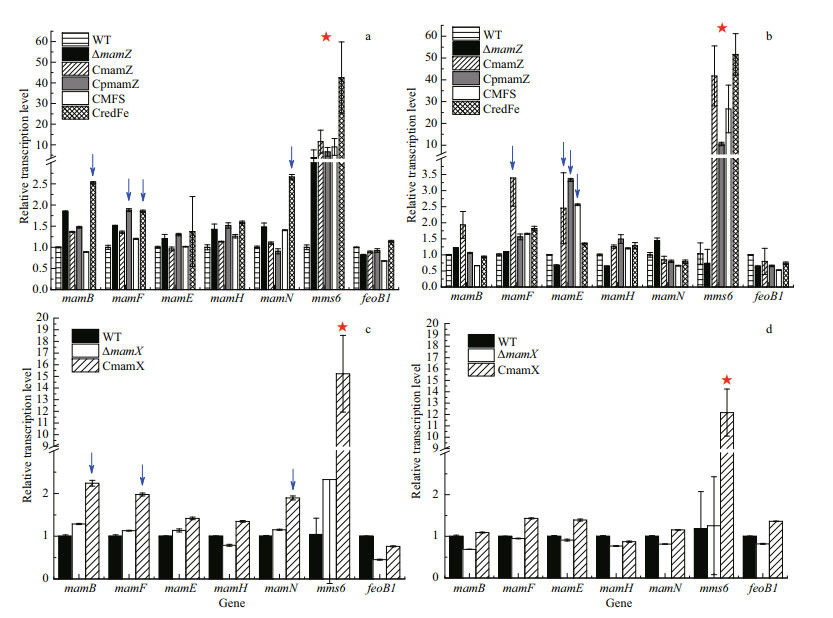
|
| Fig.6 Relative transcription levels of genes in MAI operons other than the mamXY operon a & c. 12 h; b & d. 18 h. Blue arrows: 2-fold upregulation of transcription levels. Red stars: transcription levels of mms6 were upregulated primarily in the complemented strains. |
mamZ deletion caused a dramatic increase in mms6 transcription during the logarithmic phase of cell growth (12 h). In the complemented strains, maximal levels of mms6 transcription were observed during the stationary phase (18 h), coinciding with the gradual maturation stage of magnetosomes. To rule out a possible effect of the plasmid itself, a control strain containing an empty plasmid was subjected to RT-qPCR analysis. mms6 was not upregulated at either logarithmic phase or stationary phase (Supplementary Fig.S4). Our findings suggested that exogenous fragments caused significant upregulation of mms6, and that some type of interaction occurred among the fragments. We propose that MamXY interacts with Mms6 during the magnetosome maturation process.
4 DISCUSSION 4.1 MamZ proteins function primarily during the magnetosome maturation process in MSR-1 cellsBased on the present and previous findings, we hypothesize that the four proteins encoded by genes of the mamXY operon play important roles in the magnetosome maturation process (corresponding to the stationary phase of cell growth). To elucidate the function of the highly conserved MamZ protein in MSR-1, we constructed a mutant mamZ deletion strain (ΔmamZ), firstly. mamZ deletion had no effect on growth curve, but notably reduced magnetic response.
ΔmamZ cells showed very weak magnetism of immature magnetosomes. Magnetosomes in ΔmamZ cells were small, irregular, and consisted of poor-lattice crystals, as demonstrated by HR-TEM images, similar to those of mamX, ftsZ-like, and integrated mamXY operon deletion mutant strains (Ding et al., 2010; Murat et al., 2010; Yang et al., 2013), consistent with findings by Schüler's group (Raschdorf et al., 2013). Besides, Yang et al. (2013) show that transcription level of mamZ is much higher than those of other mamXY operon genes during the period of magnetosome biosynthesis. MamZ evidently plays as important cooperative role in the magnetosome maturation process.
Bioinformatics analysis revealed two conserved domains in MamZ consisting of 18 transmembrane helices. The N-terminal domain, MFS, is homologous to MamH encoded within the mamAB operon (Raschdorf et al., 2013). The ability of MFS transporters was to move a variety of small compounds across biological membranes (Marger and Saier, 1993; Pao et al., 1998; Reddy et al., 2012). The C-terminal domain (helices belonging to FRS) shows highest similarity to YedZ in E. coli (Richter et al., 2007). YedZ is a heme B-binding membrane protein consisting of six putative transmembrane helices, while YedY is a soluble catalytic subunit in E. coli(Loschi et al., 2004; Brokx et al., 2005). YedY anchored to YedZ is a highly conserved bacterial heme-molybdoenzyme of the sulfite oxidase class found in a variety of bacteria. The importance of YedZ-like C-terminal domain in MamZ was demonstrated by Schüler's group, who deleted the YedZ-like domain and observed magnetosomes in resulting strain mamZ Δ438–639 similar to those in ΔmamZ (deletion of full-length mamZ) (Raschdorf et al., 2013). Therefore, the N-terminal MFS domain was focused on in this study. Besides, sites in MamZ for interaction with other DNAs, RNAs, and proteins mainly scatter in 281–648 aa region. Four complemented strains with differing mamZ fragment lengths (CmamZ, CpmamZ, CMFS, and CredFe) were constructed. Results from room-temperature hysteresis loops and FORC showed that the magnetism of cells was strengthen when mamZ, whether full-length or segmental, expressed in ΔmamZ. However, Cmag values of cells were still zero as long as partial mamZ was expressed in ΔmamZ. These suggested that physical properties of magnetosomes might be changed (e.g., composition, structure) (Ding et al., 2010; Murat et al., 2010; Yang et al., 2013) and various domains in MamZ might play indispensable roles in magnetosome maturation process. In iron uptake study, strains showed various iron uptake capability from medium during 0–24 h in a declining order of CmamZ, CMFS, CpmamZ, and CredFe (Supplementary Fig.S3). The difference implied the importance role of MFS in MamZ to transport iron, consistent with the analysis of bioinformation. Compared to CmamZ, CMFS, and CredFe, CpmamZ exhibited a mid-state in all of phenotypic and physiological experiments. These might be related to interaction sites scattering in partial MFS and integrated FRS. Multiple transmembrane helices and low transcription levels of MamZ could be other reasons leading to complemented strains, even CmamZ, lack the capacity for normal magnetosome maturation, and instead produce large numbers of immature particles.
4.2 Relationships (interactions) among mamZ and other key MAI genesWe determined relative transcription levels of important MAI genes by RT-qPCR. Deletion of mamZ resulted in upregulation of three other genes in the mamXY operon, indicating a close relationship among the four encoded proteins (Yang et al., 2013). In view of the overlapping region between mamZ and mamX gene, we evaluated transcription levels of MAI genes in the corresponding deletion mutants (ΔmamZ, ΔmamX) and their complemented strains, in comparison to WT. Of these genes, only mms6 showed greatly increased transcription; particularly, a 10- to 50-fold increase in the complemented strains. Previous studies have shown that Mms6 protein has a hydrophobic N-terminal region involving an interlaced structure of intra- and inter-molecular interactions, and a hydrophilic C-terminal region including an iron-binding site that plays a major role in regulation of magnetite crystal size and morphology (Arakaki et al., 2003, 2010, 2014; Amemiya et al., 2007; Feng et al., 2013). Mms6 was proposed as a specific magnetite nucleation protein and the key features for self-assembly to display a charged surface for specific iron binding, with the curvature of the surfaces determining the particle size (Staniland and Rawlings, 2016). The C-terminal region of MamZ is predicted to be a ferric-reduction domain with high similarity to YedZ in E. coli, including homologous conserved histidyl residues in the transmembrane domains that function in heme binding (von Rozycki et al., 2004). MamX contains two CXXCH heme-binding motifs typical of c-type cytochromes, which may function in electron transport or as redox buffers during magnetite formation (Siponen et al., 2012; Yang et al., 2013). Deletion of mamZ, mamX, or mms6 resulted in reduced magnetosome size (Amemiya et al., 2007; Tanaka et al., 2011; Raschdorf et al., 2013; Yang et al., 2013; Kashyap et al., 2014).
We hypothesize that MamX and MamZ proteins, in addition to having a close complementary or cooperative relationship with each other, are associated with Mms6, a protein that plays an important role in promotion of magnetosome biosynthesis both in vitro and in vivo. The several proteins encoded by MAI genes cooperate closely with each other and are all involved in magnetosome biosynthesis and maturation; none of them can be omitted. The proteins encoded by mamXY operon genes are closely related to Mms6, which was also supported by Wang et al. (2019). It indicates that Mms6 is also essential to the magnetosome biosynthesis process. Due to the presence of multiple transmembrane domains, heterologous expression of the MamZ protein is quite difficult and we are trying to perform analysis of protein interaction in vitro. In addition, most of the current studies on the function of key proteins (such as Mms6) have been conducted in vitro (Nguyen et al., 2016; Peigneux et al., 2019; Rawlings et al., 2020), and our next experiments aim at intracellular analysis of the relationship between MamZ, Mms6, and several key proteins. Moreover, the MamZ structure includes numerous binding sites for proteins and nucleic acids (both DNA and RNA), indicating that this protein interacts with such macromolecules. Studies are in progress to test this idea using a new experimental strategy for in vivo analysis.
5 CONCLUSIONIn this study, a mamZ deleted mutant Magnetospirillum gryphiswaldense MSR-1 (∆mamZ)was obtained successfully. Phenotypic and physiological experiments proved that MamZ evidently plays an important role in the magnetosome maturation process. The results of transcriptional levels by RT-qPCR showed the close relationship between mamZ and other genes in mamXY operon during the maturation process of magnetosome, especially the interaction between Mms6 and MamXY worthy for further attention.
6 DATA AVAILABILITY STATEMENTAll the data supporting the results of this study are available within the article and the supplementary material.
7 ACKNOWLEDGMENTThe authors are grateful to Dr. Yinzhao WANG (Institute of Geology and Geophysics, Chinese Academy of Sciences) for technical help, and to Dr. S. ANDERSON for English editing of the manuscript.
Electronic supplementary material Supplementary material (Supplementary Tables S1–S2 and Figs.S1–S4) is available in the online version of this article at https://doi.org/10.1007/s00343-021-0321-9.
Amemiya Y, Arakaki A, Staniland S S, Tanaka T, Matsunaga T. 2007. Controlled formation of magnetite crystal by partial oxidation of ferrous hydroxide in the presence of recombinant magnetotactic bacterial protein Mms6. Biomaterials, 28(35): 5381-5389.
DOI:10.1016/j.biomaterials.2007.07.051 |
Arakaki A, Masuda F, Amemiya Y, Tanaka T, Matsunaga T. 2010. Control of the morphology and size of magnetite particles with peptides mimicking the Mms6 protein from magnetotactic bacteria. Journal of Colloid and Interface Science, 343(1): 65-70.
DOI:10.1016/j.jcis.2009.11.043 |
Arakaki A, Webb J, Matsunaga T. 2003. A novel protein tightly bound to bacterial magnetic particles in Magnetospirillum magneticum strain AMB-1. Journal of Biological Chemistry, 278(10): 8745-8750.
DOI:10.1074/jbc.M211729200 |
Arakaki A, Yamagishi A, Fukuyo A, Tanaka M, Matsunaga T. 2014. Co-ordinated functions of Mms proteins define the surface structure of cubo-octahedral magnetite crystals in magnetotactic bacteria. Molecular Microbiology, 93(3): 554-567.
DOI:10.1111/mmi.12683 |
Balkwill D L, Maratea D, Blakemore R P. 1980. Ultrastructure of a magnetotactic spirillum. Journal of Bacteriology, 141(3): 1399-1408.
DOI:10.1128/jb.141.3.1399-1408.1980 |
Brokx S J, Rothery R A, Zhang G J, Ng D P, Weiner J H. 2005. Characterization of an Escherichia coli sulfite oxidase homologue reveals the role of a conserved active site cysteine in assembly and function. Biochemistry, 44(30): 10339-10348.
DOI:10.1021/bi050621a |
Ding Y, Li J H, Liu J N, Yang J, Jiang W, Tian J S, Li Y, Pan Y X, Li J L. 2010. Deletion of the ftsZ-like gene results in the production of superparamagnetic magnetite magnetosomes in Magnetospirillum gryphiswaldense. Journal of Bacteriology, 192(4): 1097-1105.
DOI:10.1128/JB.01292-09 |
Drew D, Slotboom D J, Friso G, Reda T, Genevaux P, Rapp M, Meindl-Beinker N M, Lambert W, Lerch M, Daley D O, van Wijk K J, Hirst J, Kunji E, de Gier J W. 2005. A scalable, GFP-based pipeline for membrane protein overexpression screening and purification. Protein Science, 14(8): 2011-2017.
DOI:10.1110/ps.051466205 |
Feng S R, Wang L J, Palo P, Liu X P, Mallapragada S K, Nilsen-Hamilton M. 2013. Integrated self-assembly of the Mms6 magnetosome protein to form an iron-responsive structure. International Journal of Molecular Sciences, 14(7): 14594-14606.
DOI:10.3390/ijms140714594 |
Frankel R B, Blakemore R P, Wolfe R S. 1979. Magnetite in freshwater magnetotactic bacteria. Science, 203(4387): 1355-1356.
DOI:10.1126/science.203.4387.1355 |
Gorby Y A, Beveridge T J, Blakemore R P. 1988. Characterization of the bacterial magnetosome membrane. Journal of Bacteriology, 170(2): 834-841.
DOI:10.1128/jb.170.2.834-841.1988 |
Heywood B R, Bazylinski D A, Garratt-Reed A, Mann S, Frankel R B. 1990. Controlled biosynthesis of greigite(Fe3S4) in magnetotactic bacteria. Naturwissenschaften, 77(11): 536-538.
DOI:10.1007/BF01139266 |
Kashyap S, Woehl T J, Liu X P, Mallapragada S K, Prozorov T. 2014. Nucleation of iron oxide nanoparticles mediated by Mms6 protein in situ. ACS Nano, 8(9): 9097-9106.
DOI:10.1021/nn502551y |
Keen N T, Tamaki S, Kobayashi D, Trollinger D. 1988. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene, 70(1): 191-197.
DOI:10.1016/0378-1119(88)90117-5 |
Kolinko I, Lohße A, Borg S, Raschdorf O, Jogler C, Tu Q, Pósfai M, Tompa E, Plitzko J M, Brachmann A, Wanner G, Müller R, Zhang Y M, Schüler D. 2014. Biosynthesis of magnetic nanostructures in a foreign organism by transfer of bacterial magnetosome gene clusters. Nature Nanotechnology, 9(3): 193-197.
DOI:10.1038/nnano.2014.13 |
Lohße A, Ullrich S, Katzmann E, Borg S, Wanner G, Richter M, Voigt B, Schweder T, Schüler D. 2011. Functional analysis of the magnetosome island in Magnetospirillum gryphiswaldense: the mamAB operon is sufficient for magnetite biomineralization. PLoS One, 6(10): e25561.
DOI:10.1371/journal.pone.0025561 |
Loschi L, Brokx S J, Hill T L, Zhang G, Bertero M G, Lovering A L, Weiner J H, Strynadka N C J. 2004. Structural and biochemical identification of a novel bacterial oxidoreductase. Journal of Biological Chemistry, 279(48): 50391-50400.
DOI:10.1074/jbc.M408876200 |
Madej M G, Dang S Y, Yan N E, Kaback H R. 2013. Evolutionary mix-and-match with MFS transporters. Proceedings of the National Academy of Sciences of the United States of America, 110(15): 5870-5874.
DOI:10.1073/pnas.1303538110 |
Madej M G, Kaback H R. 2013. Evolutionary mix-and-match with MFS transporters II. Proceedings of the National Academy of Sciences of the United States of America, 110(50): e4831-e4838.
DOI:10.1073/pnas.1319754110 |
Marger M D, Saier Jr M H. 1993. A major superfamily of transmembrane facilitators that catalyse uniport, symport and antiport. Trends in Biochemical Sciences, 18(1): 13-20.
DOI:10.1016/0968-0004(93)90081-W |
Murat D, Quinlan A, Vali H, Komeili A. 2010. Comprehensive genetic dissection of the magnetosome gene island reveals the step-wise assembly of a prokaryotic organelle. Proceedings of the National Academy of Sciences of the United States of America, 107(12): 5593-5598.
DOI:10.1073/pnas.0914439107 |
Nguyen H V, Suzuki E, Oestreicher Z, Minamide H, Endoh H, Fukumori Y, Taoka A. 2016. A protein-protein interaction in magnetosomes: TPR protein MamA interacts with an Mms6 protein. Biochemistry and Biophysics Reports, 7: 39-44.
DOI:10.1016/j.bbrep.2016.05.010 |
Nudelman H, Zarivach R. 2014. Structure prediction of magnetosome-associated proteins. Frontiers in Microbiology, 5: 9.
DOI:10.3389/fmicb.2014.00009 |
Pao S S, Paulsen I T, Saier Jr M H. 1998. Major facilitator superfamily. Microbiology and Molecular Biology Reviews, 62(1): 1-34.
DOI:10.1128/MMBR.62.1.1-34.1998 |
Peigneux A, Jabalera Y, Vivas M A F, Casares S, Azuaga A I, Jimenez-Lopez C. 2019. Tuning properties of biomimetic magnetic nanoparticles by combining magnetosome associated proteins. Scientific Reports, 9: 8804.
DOI:10.1038/s41598-019-45219-7 |
Raschdorf O, Müller F D, Pósfai M, Plitzko J M, Schüler D. 2013. The magnetosome proteins MamX, MamZ and MamH are involved in redox control of magnetite biomineralization in Magnetospirillum gryphiswaldense. Molecular Microbiology, 89(5): 872-886.
DOI:10.1111/mmi.12317 |
Rawlings A E, Liravi P, Corbett S, Holehouse A S, Staniland S S. 2020. Investigating the ferric ion binding site of magnetite biomineralisation protein Mms6. PLoS One, 15(2): e0228708.
DOI:10.1371/journal.pone.0228708 |
Reddy V S, Shlykov M A, Castillo R, Sun E I, Saier Jr M H. 2012. The major facilitator superfamily (MFS) revisited. FEBS Journal, 279(11): 2022-2035.
DOI:10.1111/j.1742-4658.2012.08588.x |
Richter M, Kube M, Bazylinski D A, Lombardot T, Glöckner F O, Reinhardt R, Schüler D. 2007. Comparative genome analysis of four magnetotactic bacteria reveals a complex set of group-specific genes implicated in magnetosome biomineralization and function. Journal of Bacteriology, 189(13): 4899-4910.
DOI:10.1128/JB.00119-07 |
Rong C B, Huang Y J, Zhang W J, Jiang W, Li Y, Li J L. 2008. Ferrous iron transport protein B gene (feoB1) plays an accessory role in magnetosome formation in Magnetospirillum gryphiswaldense strain MSR-1. Research in Microbiology, 159(7-8): 530-536.
DOI:10.1016/j.resmic.2008.06.005 |
Sakaguchi T, Burgess J G, Matsunaga T. 1993. Magnetite formation by a sulphate-reducing bacterium. Nature, 365(6441): 47-49.
DOI:10.1038/365047a0 |
Schüler D. 2004. Molecular analysis of a subcellular compartment: the magnetosome membrane in Magnetospirillum gryphiswaldense. Archives of Microbiology, 181(1): 1-7.
DOI:10.1007/s00203-003-0631-7 |
Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nature Biotechnology, 1(9): 784-791.
DOI:10.1038/nbt1183-784 |
Siponen M I, Adryanczyk G, Ginet N, Arnoux P, Pignol D. 2012. Magnetochrome: a c-type cytochrome domain specific to magnetotatic bacteria. Biochemical Society Transactions, 40(6): 1319-1323.
DOI:10.1042/BST20120104 |
Staniland S S, Rawlings A E. 2016. Crystallizing the function of the magnetosome membrane mineralization protein Mms6. Biochemical Society Transactions, 44(3): 883-890.
DOI:10.1042/BST20160057 |
Tanaka M, Arakaki A, Matsunaga T. 2010. Identification and functional characterization of liposome tubulation protein from magnetotactic bacteria. Molecular Microbiology, 76(2): 480-488.
DOI:10.1111/j.1365-2958.2010.07117.x |
Tanaka M, Mazuyama E, Arakaki A, Matsunaga T. 2011. Mms6 protein regulates crystal morphology during nanosized magnetite biomineralization in vivo. Journal of Biological Chemistry, 286(8): 6386-6392.
DOI:10.1074/jbc.M110.183434 |
von Rozycki T, Yen M R, Lende E E, Saier Jr M H. 2004. The YedZ family: possible heme binding proteins that can be fused to transporters and electron carriers. Journal of Molecular Microbiology and Biotechnology, 8(3): 129-140.
DOI:10.1159/000085786 |
Wang Q, Liu J X, Zhang W J, Zhang T W, Yang J, Li Y. 2013. Expression patterns of key iron and oxygen metabolism genes during magnetosome formation in Magnetospirillum gryphiswaldense MSR-1. FEMS Microbiology Letters, 347(2): 163-172.
DOI:10.1111/1574-6968.12234 |
Wang Q, Wang M W, Wang X, Guan G H, Li Y, Peng Y L, Li J L. 2015a. Iron response regulator protein IrrB in Magnetospirillum gryphiswaldense MSR-1 helps control the iron/oxygen balance, oxidative stress tolerance, and magnetosome formation. Applied and Environmental Microbiology, 81(23): 8044-8053.
DOI:10.1128/AEM.02585-15 |
Wang Q, Wu S, Li X Y, Zhang T W, Yang J, Wang X, Li F, Li Y, Peng Y L, Li J L. 2019. Work patterns of MamXY proteins during magnetosome formation in Magnetospirillum gryphiswaldense MSR-1. Applied and Environmental Microbiology, 85(2): e02394-18.
DOI:10.1128/AEM.02394-18 |
Wang X, Wang Q, Zhang W J, Wang Y J, Li L, Wen T, Zhang T W, Zhang Y, Xu J, Hu J Y, Li S Q, Liu L Z, Liu J X, Jiang W, Tian J S, Li Y, Schüler D, Wang L, Li J L. 2014. Complete genome sequence of Magnetospirillum gryphiswaldense MSR-1. Genome Announcements, 2(2): e00171-14.
DOI:10.1128/genomeA.00171-14 |
Wang Y Z, Lin W, Li J H, Zhang T W, Li Y, Tian J S, Gu L X, Heyden Y V, Pan Y X. 2015b. Characterizing and optimizing magnetosome production of Magnetospirillum sp.XM-1 isolated from Xi'an City Moat, China. FEMS Microbiology Letters, 362(21): fnv167.
DOI:10.1093/femsle/fnv167 |
Yan N E. 2013. Structural advances for the major facilitator superfamily (MFS) transporters. Trends in Biochemical Sciences, 38(3): 151-159.
DOI:10.1016/j.tibs.2013.01.003 |
Yang J, Li S Q, Huang X L, Li J H, Li L, Pan Y X, Li Y. 2013. MamX encoded by the mamXY operon is involved in control of magnetosome maturation in Magnetospirillum gryphiswaldense MSR-1. BMC Microbiology, 13: 203.
DOI:10.1186/1471-2180-13-203 |
Zhang C, Meng X, Li N X, Wang W, Sun Y, Jiang W, Guan G H, Li Y. 2013. Two bifunctional enzymes with ferric reduction ability play complementary roles during magnetosome synthesis in Magnetospirillum gryphiswaldense MSR-1. Journal of Bacteriology, 195(4): 876-885.
DOI:10.1128/JB.01750-12 |
 2021, Vol. 39
2021, Vol. 39



