Institute of Oceanology, Chinese Academy of Sciences
Article Information
- CHEN Haitao, LI Jinhua, WU Long-Fei, ZHANG Wei-Jia
- Morphological and phylogenetic diversity of magnetotactic bacteria and multicellular magnetotactic prokaryotes from a mangrove ecosystem in the Sanya River, South China
- Journal of Oceanology and Limnology, 39(6): 2015-2026
- http://dx.doi.org/10.1007/s00343-021-0491-5
Article History
- Received Dec. 30, 2020
- accepted in principle May. 6, 2021
- accepted for publication Jun. 9, 2021
2 France-China Joint Laboratory for Evolution and Development of Magnetotactic Multicellular Organisms, Chinese Academy of Sciences, Beijing 100029, China;
3 Deep-Sea Microbial Cell Biology, Department of Deep Sea Sciences, Institute of Deep-Sea Science and Engineering, Chinese Academy of Sciences, Sanya 572000, China;
4 Key Laboratory of Earth and Planetary Physics, Institute of Geology and Geophysics, Chinese Academy of Sciences, Beijing 100029, China;
5 LCB, Aix Marseille University, CNRS, Marseille 13402, France
Magnetotactic bacteria (MTB) are a group of morphologically and phylogenetically heterogeneous prokaryotes. They are capable of sensing and changing their orientation in accordance with the direction of the geomagnetic field, a behavior called magnetotaxis (Faivre and Schüler, 2008; Komeili, 2012). This unique capability is facilitated by special organelles that are intracellularly synthesized, membrane-enclosed ferromagnetic nanocrystals of magnetite (Fe3O4) and/or greigite (Fe3S4), called magnetosomes (Bazylinski and Frankel, 2004). Because of the special properties of these magnetosomes, MTB have been exploited for applications in diverse disciplines from geobiology to biotechnology (Komeili, 2012; Li et al., 2013). In addition, the fossil magnetosomes preserved in sediments (called magnetofossils) could be used as potential proxies for reconstructing the paleoenvironment and paleo-climate changes and understanding the origin and evolution of early life (Pan et al., 2005; Kopp and Kirschvink, 2008; Li et al., 2020b).
MTB are widely distributed in various aquatic environments such as lakes, rivers, ponds, estuaries, lagoons, marine sediments, and even deep-sea extreme environments (Petersen et al., 1986; Lins et al., 2007; Jogler et al., 2010; Chen et al., 2015a; Li et al., 2016; Lin et al., 2017, 2018; Liu et al., 2017). Different morphologies have been described, including single cells in forms of cocci, ovoid, bean-shaped, rod-shaped, and spirilla, and groups of cell aggregates composing of up to over forty ovoid cells, which are known as multicellular magnetotactic prokaryotes (MMPs) (Abreu et al., 2007; Lins et al., 2007; Isambert et al., 2007; Pan et al., 2008; Lin et al., 2009; Wenter et al., 2009; Zhou et al., 2011; Qian et al., 2020). Phylogenetically, most known MTB are affiliated with the phyla Proteobacteria, Nitrospirae, Planctomycetes, the candidate phyla "Omnitrophica" and "Latescibacteria" (Lefèvre et al., 2009; Lefèvre and Bazylinski, 2013; Lin and Pan, 2015; Dziuba et al., 2016; Lin et al., 2017; Liu et al., 2021). Despite of the abundant data regarding cellular and crystal characterization, and phylogenetic diversity of MTB (Lins et al., 2000; Zhou et al., 2011; Wang et al., 2012; Chen et al., 2015b; Lin et al., 2017), only a few MTB strains have been successfully isolated and cultivated and the physiological characters of certain phylogenetic group of MTB remain obscure (Schüler and Köhler, 1992; Nakamura et al., 1993; Lefèvre et al., 2009, 2011; Schübbe et al., 2009; Zhu et al., 2010; Wang et al., 2013). Recently, fluorescence in-situ hybridization coupled with scanning (FISH-SEM) or transmission electron microscopy (FISH-TEM) was developed (Li et al., 2017). It enabled distinguishing MTB from other microorganisms in environmental samples at the single-cell level, and connecting phylogeny with the cell ultrastructure and magnetosome morphology of uncultured MTB strains (Li et al., 2017, 2019; Zhang et al., 2017; Koziaeva et al., 2020; Qian et al., 2020; Liu et al., 2021).
Previous studies have shown that diversity and communities of MTB, as well as the magnetic crystal morphology and spatial arrangement of magnetosomes, varied depending on environmental factors, such as the strength of the earth's magnetic field, geographic latitude, temperature, salinity, pH, and nutrients (Lefèvre and Wu, 2013; Pósfai et al., 2013; Lin et al., 2014; Li et al., 2020a, b ; Liu et al., 2021). Mangroves are peculiar ecosystems connecting terrestrial and marine environments in tropical and subtropical regions. The woody plants growing in coastal wetlands are periodically flooded by tidal water, which possibly result in a diverse and dynamic microbial community (Alongi, 2002). In general, study of MTB in mangrove ecosystem is lacked, with only limited reports of the presence of magnetite magnetic minerals and coccoid MTB in mangroves samples (Kannapiran et al., 1999; Maloof et al., 2007; Lin et al., 2012). To better understand the phylogenetic diversity and characterization of MTB in mangroves, we collected MTB from mangrove estuary waters in Sanya, southern China. Optical and electronic microscopes were used to analyze their abundance and morphology. Taxonomic identification and phylogeny study of collected cells were carried out by analyzing the 16S rRNA gene sequences. Additionally, we characterized the morphology of bacterial cells and magnetosomes of certain phylogenetic group of MTB with FISH-TEM analysis.
2 MATERIAL AND METHOD 2.1 Sampling and magnetic collectionSediment samples were collected from the tidal flats of mangrove located at the mouth of the Sanya River (Sanya, China; 18°15.242ʹN, 109°30.585ʹE) in spring of 2014 and 2015. To carry out enrichment of magnetotactic bacteria, approximately 300-mL sediments were transferred to 0.5-L plastic bottles covered with ~200 mL of seawater collected from the surface layer (~3–10 cm) at the sampling sites. A piece of magnet was stick to the bottle wall approximately 2 cm below the interface between the sediment and surface water with the southern pole facing interior. The bottles were covered loosely and stored at room temperature under dim light for a week for the enrichment. The pH, temperature, and salinity of sediments measured in-situ were 8.2, 23 ℃, and 21.33, respectively. The concentrations of NH4+, NO2-, NO3-, SO2-, and PO3- in pore water were determined by using Elemental analyzer (Hach DR890, USA) to be 8.4, 0.02, 1.23, 2017, and 7.3 mg/L, respectively. Collection of MTB was purified by race-track method (Wolfe et al., 1987; Li et al., 2010).
2.2 Optical microscopyThe quantity and community of MTB in the microcosms were periodically checked with hanging drop method using an optical microscope at a 40× magnification (Olympus BX53, Japan). Based on these microscopic images, cell size of MMPs was measured with software ImageJ 1.51i.
2.3 Transmission electron microscopy (TEM)The purified sample was deposited on a carbon-coated copper grid supported by formvar for TEM observation. A JEM-2100 TEM equipped with an Oxford SDD detector (X-MaxN 80T) and an accelerating voltage of 200 kV was used for morphological and cellular structure studies. Under the same conditions, the chemical composition and structure of magnetic crystals were analyzed by energy dispersive X-ray spectroscopy (EDXS), and high-resolution TEM (HRTEM). The crystal length (along the long axis) and width (perpendicular to the long axis) of the magnetosomes were measured from the TEM images by software ImageJ 1.51i. Three MMPs cells of each morphological type were analyzed for the characterization of the magnetosome crystals.
2.4 PCR amplification, 16S rRNA gene sequencing, and phylogenetic analyses16S rRNA gene sequences of the samples were amplified as described previously by Chen et al. (2015a). With bacteria-specific primers 27F and 1492R, PCR reactions were performed using a T100TM Thermal Cycler (Bio-Rad, America). The cycling conditions were 4 min at 94 ℃ for initial denaturation; 30 cycles of 1 min at 94 ℃, 45 s at 55 ℃, and 1.5 min at 72 ℃; and a final extension of 10 min at 72 ℃. PCR products were purified, ligated into the pMD19-T cloning vector (TaKaRa, Japan) and transformed into Escherichia coli DH5α to construct a 16S rRNA gene clone library. Positive clones were confirmed by PCR amplification with primer M-13 and RV-M. 60 clones were sequenced by Shanghai Invitrogen Biotechnology Co., Ltd. Using BLAST search program on the NCBI website, the 16S rRNA gene sequences retrieved in this study were first analyzed. The related sequences were then aligned with their close relatives using the ClustalW algorithm, and a phylogenetic tree was subsequently constructed using the neighbor-joining method in MEGA version 6.0 (Kumar et al., 2016). The sequences were submitted in GenBank database with accession numbers MW356763-MW356778.
2.5 Coupled FISH-TEM analysisIn this study, the Alphaproteobacteria-specific probe ALF968 was used for fluorescence in-situ hybridization (Neef, 1997). The bacterial universal probe EUB338 was used as a control (Amann et al., 1990). The probe ALF968 was labeled at the 5' end with hydrophilic sulfoindocyanine Cy3, and the universal probe was labeled at the 5' end with fluoresce in phosphoramidite (FAM). Both probes were synthesized and fluorescently labeled by Invitrogen Co., Ltd. Coupled FISH-TEM analysis was based on the procedure described in Li et al. (2017). The collected MTB cells were fixed with 4% paraformaldehyde as described by Pernthaler et al. (2002) and then washed in 1×phosphate buffer saline. By centrifugation, the cells were fixed (dehydrated) for the second time with 50%, 80%, and 100% ethanol, respectively. Then, the cells were resuspended in hybridization buffer containing probe (50 ng/μL) for in-situ hybridization at 46 ℃ for 3 h. Afterwards, the samples were washed three times with elution buffer and water respectively. An amount of 5-μL washed cells was deposited onto letter-indexed carbon-coated copper grids, dried in air at ambient temperature. The results were recorded using a fluorescence microscope (Olympus BX51) and TEM (JEM-2100 TEM).
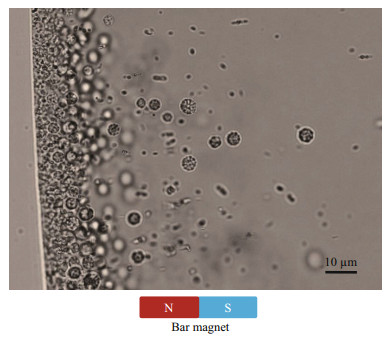
3 RESULT AND DISCUSSION 3.1 Morphologic and chemical features of MTB from Sanya mangroveLiving MTB collected from sediment samples were observed with optical microscopy. The cells abundance was approximately 103 cells/cm3. The MTB collected from Sanya mangrove were morphologically diverse and consisted of both unicellular MTB and multicellular MMPs. The multicellular MMPs represented as an abundant population in the samples collected. They exhibited a spherical morphology (s-MMPs) with a diameter of 4.6±0.2 μm (Fig. 1). Like s-MMPs observed in other marine environments, they showed classic escape motility or "ping-pong" motion that undergoes backward excursions followed by a forward movement (Lins et al., 2007; Wenter et al., 2009; Zhou et al., 2011).
|
| Fig.1 MTB collected from the mangrove of the Sanya River MTB swam northwards and accumulated at the edge of the droplet. Because of the escape motion, s-MMPs swam away southward from the edge, which made them easier for observation compared to other MTB. |
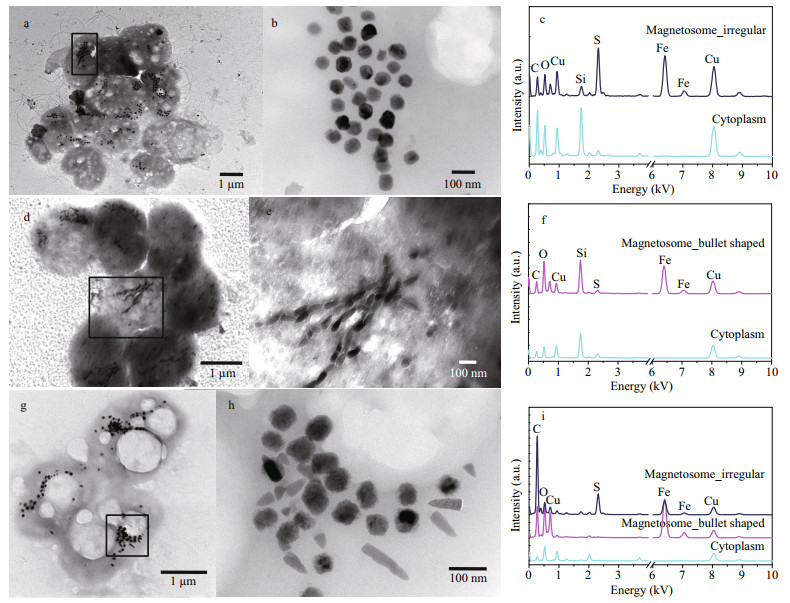
The s-MMPs could be divided into three different morphotypes in shape, size, and composition of magnetosomes (Fig. 2 & Table 1). Type-I likely contains only greigite-type magnetosomes on average size of 77±11 nm in length (the number of magnetosomes: n=263; Table 1). Type-Ⅱ likely contains only bullet shaped magnetite magnetosomes on average size of 78±18 nm in length and 34±4 nm in width (n=125). Type-Ⅲ contains both magnetite-type and greigite-type magnetosomes, the average sizes of magnetosomes are 80±19 nm for greigite crystals, and 88±19 nm in length and 34±5 nm in width (n=110) for bullet-shaped magnetite crystals (Fig. 2g–i). The three different types of s-MMPs were also reported in the lagoons of Brazil (Lins et al., 2007). Comparatively, the average size of magnetosomes formed by the Type-I was slightly larger than those observed in the lagoons in Brazil (70±8 nm) and Yuehu Lake (63.9±9.3 nm) (Lins et al., 2007; Zhang et al., 2014). The average size of bullet-shaped magnetosomes produced by Type-Ⅱ was smaller than those in previously reported s-MMPs collected elsewhere (Lins et al., 2007; Wenter et al., 2009; Zhou et al., 2011, 2013; Zhang et al., 2014). To summarize, the s-MMPs discovered from the mangrove sediments of the Sanya River resembled s-MMPs in other ecosystems in cell size (Table 1) and movement behavior, but differed slightly in the morphology of magnetosomes (Lins et al., 2007; Wenter et al., 2009; Zhou et al., 2011). This agrees with current understanding that magnetosome biomineralization is biologically controlled, yet affected by environmental factors of habitats at certain degree (Li and Pan, 2012).
|
| Fig.2 Morphological and chemical features of MMPs collected from Sanya mangrove environment a–c. TEM images of Type-I MMP of cell (a) and magnetosomes (b), and the corresponding EDXS microanalysis of magnetosomes (c), respectively; d–f. TEM images of Type-Ⅱ MMP of cell (d) and magnetosomes (e), and the corresponding EDXS microanalysis of magnetosomes, respectively; g–i. TEM images of Type-Ⅲ MMP of cell (g) and magnetosomes (h), and the corresponding EDXS microanalysis of magnetosomes (i). In (c), (f), and (i), the light blue, pink, and blue lines indicate the EDXS signals collected from a small area (~50 nm) in cell wall, magnetite-type magnetosomes, and greigite-type magnetosomes, respectively. |
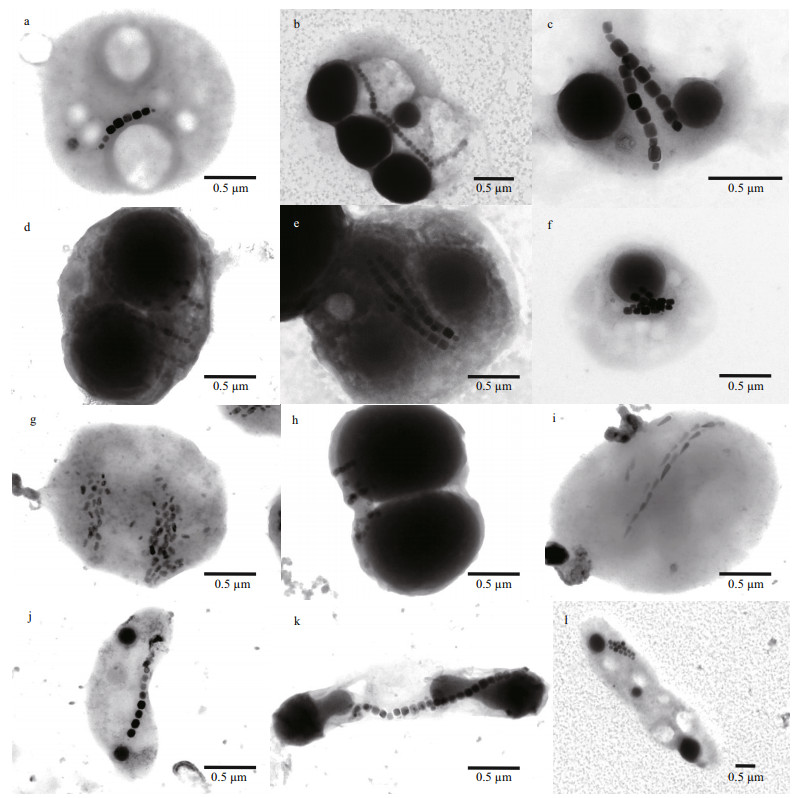
As expected, TEM observations revealed a highly diverse unicellular MTB in terms of cellular and magnetic crystal morphologies. As shown in Fig. 3, we found elongated prismatic magnetosomes in cocci (Fig. 3a–h), vibrio (Fig. 3j), and rod-shaped bacteria (Fig. 3k–l); and bullet-shaped magnetosomes in magnetotactic cocci (Fig. 3i). The magnetosomes are arranged in single chains (Fig. 3a–b & j–k), double chains (Fig. 3c, i, & l), multiple chains (Fig. 3d, e, f, & g), or in scattered distribution (Fig. 3h). Consistent with previous studies (Spring et al., 1993; Cox et al., 2002; Bazylinski et al., 2004; Lefèvre et al., 2011; Li et al., 2017, 2019, 2020a; Rivas-Lamelo et al., 2017; Schulz-Vogt et al., 2019), MTB collected from Sanya mangrove environments contain various micrometer-sized intracellular inclusions (e.g., sulfur-rich or polyphosphate particles, or polyhydroxyalkanoate granules) in addition to the nanometer-sized magnetosomes, indicating their involvement in driving biogeochemical cycles of key elements in nature (Li et al., 2020a).
|
| Fig.3 Transmission electron micrograph of unicellular MTB collected from Sanya mangrove environments a–i. magnetotactic cocci with prismatic (a–h) and bullet-shaped magnetosomes (i), which are organized into single chain (a, b), double chains (c, i), two double chains (e, f), non-linear chain (h), and multiple-bundled chains (d, g); j. magnetotactic vibrio with single magnetosome chain; k. rod-shaped MTB with single magnetosome chain; l. rod-shaped MTB with one double magnetosome chains. |
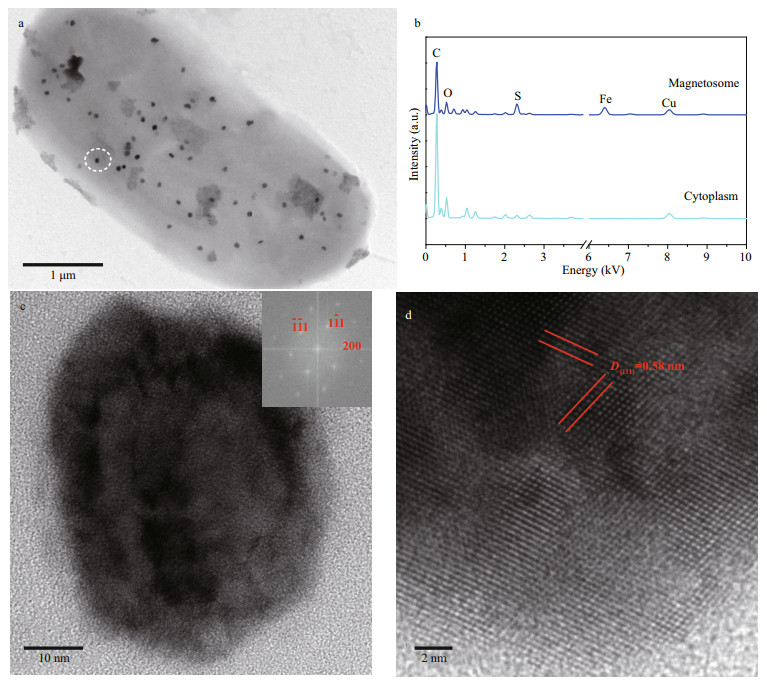
Interestingly, we found a special type of large rod-shaped bacteria whose irregularly shaped magnetosomes dispersed evenly in the cell (Fig. 4a). EDXS microanalysis showed that the magnetosomes are rich in iron and sulfur (Fig. 4b). HRTEM imaging further indicated that the mineral phase of irregular crystal was greigite (Fig. 4c–d). Large rod-shaped MTB have been reported in the Great Boiling Springs geothermal field in Gerlach, Nevada, USA, and freshwater niche previously (Lefèvre et al., 2011; Wang et al., 2013). But, in both cases, the irregularly shaped greigite magnetosomes were aligned in multiple chains. Whether the large rod-shaped MTB present in Sanya mangrove belonged to a different group, or exhibited a different magnetosome spatial arrangement due to the changed environment parameters require further investigations.
|
| Fig.4 Large rod-shaped MTB collected from Sanya mangrove environments a. TEM image; b: EDXS microanalysis of the magnetosome indicated by the white dotted circle in (a); c. HRTEM image of one individual magnetosome crystal recorded from [011] zone axis of greigite crystal. The inset is the corresponding Fast Fourier Transform pattern of the particle (c); d. the closed-up of HRTEM image of the particle (c) showing the lattice structure and the corresponding d-spacing value of plane of greigite crystal. |
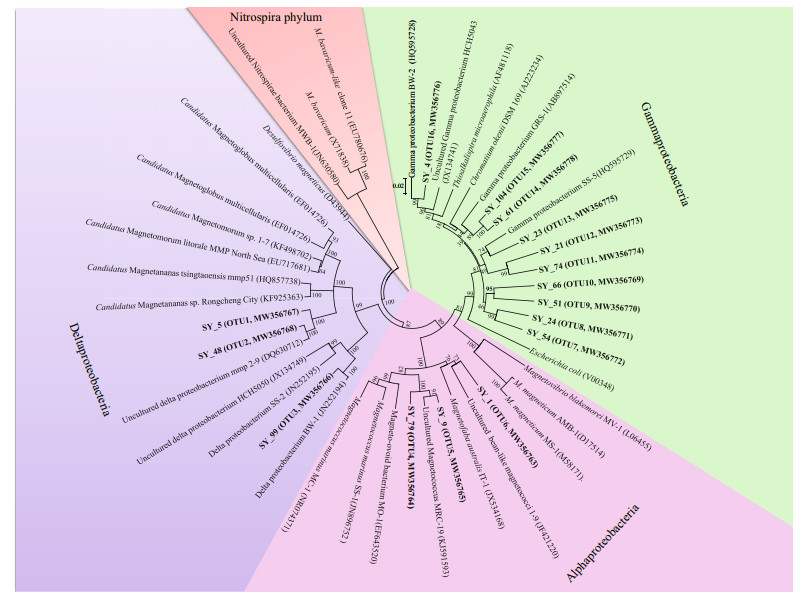
Phylogenetic analysis based on 16S rRNA gene sequences was performed to determine the community structure of MTB. In total 16 operational taxonomic units (OTUs) were identified at the 98% similarity level, all affiliated with the Proteobacteria phylum (Fig. 5). The OTU 1 and 2 shared 93.5% and 97.7% sequence identities to MMPs of the uncultured Deltaproteobacterium mmp2_9 (Simmons and Edwards, 2007), and 90.5% and 88.7% identities to the Candidatus Magnetoglobus multicellularis from Araruama Lagoon in brazil, respectively (Martins et al., 2009). Although our results showed the s-MMPs found in Sanya were similar to the MMPs of Lagoon in cell morphology and magnetosome characteristics, the relatively low similarity of 16S rRNA gene suggested that these two OTUs might represent novel MMPs species. The OTU 3 is 98.3% identical to the cultured Desulfamplus magnetovallimortis BW-1 within the Deltaproteobacteria (Lefèvre et al., 2011; Descamps et al., 2017). Three OTUs (OTU 4 to 6) belong to the Alphaproteobacteria, which were most closely related to known magnetotactic cocci (90% to 98%). Additionally, ten OTUs (OTU 7 to 16) belonged to the Gammaproteobacteria, which accounted for more than 47% of all retrieved sequences. OTU 16 was 95.6% identical to the cultured Gammaproteobacteria BW-2 (Lefèvre et al., 2012). OTUs 14 and 15 were closely related to previously described Gammaproteobacterium GRS-1 MTB (Taoka et al., 2014). OTU 13 had a maximum similarity of 90.6% with the cultured Gammaproteobacteria SHHR-1 (Zhang et al., 2017). Sequences belonging to OTU 7 to 12 were highly divergent from known MTB species and might therefore represent novel branches. Notably, the coccoid MTB identified in the saline samples from the mangrove swamps (Wenchang, China) by Lin et al. (2012) were affiliated with the uncultivated Alphaproteobacteria Magnetococcus. Comparing to that, the MTB in the mangroves of Sanya showed higher diversity in respect to morphology and phylogeny. This finding expands the knowledge of the community composition and distribution of MTB in mangrove environments, and suggests the highly adapted MTB to complex environments.
|
| Fig.5 Phylogenetic tree based on 16S rRNA gene sequences showing the relationship of the MTB obtained from the Sanya River sediment within its closest relatives The tree was constructed using neighbor-joining analysis. Bootstrap values at nodes are percentages of 1 000 replicates. The 16S rRNA gene sequence of the Nitrospirae phylum MTB was used to root the tree. GenBank accession numbers are given in parentheses. |
By the time this work was carried out, a large number of studies have shown that magnetotactic cocci belong to Alphaproteobacteria. Magnetotactic cocci with different characteristics were observed in the sediments of Sanya mangrove. Along with that, several 16S sequences retrieved from this sample belonged to the cluster containing Magnetococcus within Alphaproteobacteria. To identify the morphology and magnetosomes characteristics of Alphaproteobacteria MTB in mangrove, we carried out FISH-TEM analyses using a probe specific to the 16S rRNA of Alphaproteobacteria group (ALF968) (Neef, 1997) and a universal bacterial probe EUB338 (Amann et al., 1990) as a control. As shown in Fig. 6, the Alphaproteobacteria-specific probe ALF968 hybridized with vibrio, cocci, and rod-shaped MTB from mangrove sediment. TEM analysis showed they synthesized elongated prismatic magnetite crystals (Fig. 6c–f).

|
| Fig.6 Correlative FISH-TEM analysis of uncultured MTB from the tidal flats of mangrove located in the mouth of the Sanya River in China based on in-situ hybridization with the Alphaproteobacteria-specific probe ALF968 a–c. the same microscopic fields are shown after hybridization with the 5ʹ-FAM-labeled universal bacterial probe EUB338 (green) (a), the 5ʹ-Cy3-labeled Alphaproteobacteria-specific probe ALF968 (red) (b), and the TEM images (c). d–f. high-magnification TEM images of 1-3 in (c). |
In addition, based on 16S rRNA gene analysis we found that ten OTUs may be belonging to Gammaproteobacteria. OTU 15 shared 92.4% sequence identities to Gammaproteobacterium GRS-1 MTB. To identify whether this group is a new genus of MTB, we designed a specific probe for OTU 15, but our results showed that cells recognized by this probe did not possess magnetosomes (data not shown). We cannot rule out the possibility that this bacterium was MTB but did not synthesize magnetosomes under current condition, but it is also possible that the OTU 15 may not be MTB even though its 16S sequence shows similarity with known MTB and the accuracy of judging the diversity of MTB based on 16S rRNA gene sequences is worth discussion.
4 CONCLUSIONBoth microscopy and molecular analyses revealed a high diversity of MTB in Sanya mangrove environments. Additional to unicellular cocci, vibrios, and rod-shaped bacteria, three types of Desulfobacteraceae family belonging to Deltaproteobacterium of s-MMPs were observed: Type-I forming only irregular greigite magnetosomes, Type-Ⅱ forming only bullet-shaped magnetite magnetosomes, and Type-Ⅲ producing both bullet-shaped magnetite and irregular greigite crystals. Consistent with morphological diversity, phylogenetic analyses based on the 16S rRNA genes sequencing revealed 16 OTUs of MTB that affiliated to Alphaproteobacteria, Deltaproteobacteria, and Gammaproteobacteria. This finding suggests that the mangrove environments contain abundant and potentially novel MTBs that worth further investigations.
5 DATA AVAILABILITY STATEMENTThe data used in the paper are available from the authors.
6 ACKNOWLEDGMENTWe are grateful to Qunjian YIN for help in sample collection. The authors also thank the anonymous reviewers for valuable comments and the editors for careful editing.
Abreu F, Martins J L, Silveira T S, Keim C N, de Barros H G P L, Filho F J G, Lins U. 2007. 'Candidatus Magnetoglobus multicellularis', a multicellular, magnetotactic prokaryote from a hypersaline environment. International Journal of Systematic and Evolutionary Microbiology, 57(6): 1318-1322.
DOI:10.1099/ijs.0.64857-0 |
Alongi D M. 2002. Present state and future of the world's mangrove forests. Environmental Conservation, 29(3): 331-349.
DOI:10.1017/s0376892902000231 |
Amann R I, Krumholz L, Stahl D A. 1990. Fluorescentoligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. Journal Bacteriology, 172(2): 762-770.
DOI:10.1128/jb.172.2.762-770.1990 |
Bazylinski D A, Dean A J, Williams T J, Long L K, Middleton S L, Dubbels B L. 2004. Chemolithoautotrophy in the marine, magnetotactic bacterial strains MV-1 and MV-2. Archives of Microbiology, 182(5): 373-387.
DOI:10.1007/s00203-004-0716-y |
Bazylinski D A, Frankel R B. 2004. Magnetosome formation in prokaryotes. Nature Reviews Microbiology, 2(3): 217-230.
DOI:10.1038/nrmicro842 |
Chen H T, Li J H, Xing X, Du Z J, Chen G J. 2015a. Unexpected diversity of magnetococci in intertidal sediments of Xiaoshi Island in the North Yellow Sea. Journal of Nanomaterials, 2015: 902121.
DOI:10.1155/2015/902121 |
Chen Y R, Zhang R, Du H J, Pan H M, Zhang W Y, Zhou K, Li J H, Xiao T, Wu L F. 2015b. A novel species of ellipsoidal multicellular magnetotactic prokaryotes from Lake Yuehu in China. Environmental Microbiology, 17(3): 637-647.
DOI:10.1111/1462-2920.12480 |
Cox B L, Popa R, Bazylinski D A, Lanoil B, Douglas S, Belz A, Engler D L, Nealson K H. 2002. Organization and elemental analysis of P-, S-, and Fe-rich inclusions in a population of freshwater magnetococci. Geomicrobiology Journal, 19(4): 387-406.
DOI:10.1080/01490450290098504 |
Descamps E C T, Monteil C L, Menguy N, Ginet N, Pignol D, Bazylinski D A, Lefèvre C T. 2017. magnetovallimortis gen. nov., sp. nov., a magnetotactic bacterium from a brackish desert spring able to biomineralize greigite and magnetite, that represents a novel lineage in the Desulfobacteraceae. Systematic and Applied Microbiology, 40(5): 280-289.
DOI:10.1016/j.syapm.2017.05.001 |
Dziuba M, Koziaeva V, Grouzdev D, Burganskaya E, Baslerov R, Kolganova T, Chernyadyev A, Osipov G, Andrianova E, Gorlenko V, Kuznetsov B. 2016. Magnetospirillum caucaseum sp. nov., Magnetospirillum marisnigri sp. nov. and Magnetospirillum moscoviense sp. nov., freshwater magnetotactic bacteria isolated from three distinct geographical locations in European Russia. International Journal of Systematic and Evolutionary Microbiology, 66(5): 2069-2077.
DOI:10.1099/ijsem.0.000994 |
Faivre D, Schüler D. 2008. Magnetotactic bacteria and magnetosomes. Chemical Reviews, 108(11): 4875-4898.
DOI:10.1021/cr078258w |
Isambert A, Menguy N, Larquet E, Guyot F, Valet J P. 2007. Transmission electron microscopy study of magnetites in a freshwater population of magnetotactic bacteria. American Mineralogist, 92(4): 621-630.
DOI:10.2138/am.2007.2278 |
Jogler C, Niebler M, Lin W, Kube M, Wanner G, Kolinko S, Stief P, Beck A J, De Beer D, Petersen N, Pan Y, Amann R, Reinhardt R, Schüler D. 2010. Cultivationindependent characterization of 'Candidatus Magnetobacterium bavaricum' via ultrastructural, geochemical, ecological and metagenomic methods. Environmental Microbiology, 12(9): 2466-2478.
DOI:10.1111/j.1462-2920.2010.02220.x |
Kannapiran E, Purushothaman A, Kannan L, Saravanan S. 1999. Magneto bacteria from estuarine, mangrove and coral reef environs in Gulf of Mannar. Indian Journal of Marine Sciences, 28(3): 332-334.
|
Komeili A. 2012. Molecular mechanisms of compartmentalization and biomineralization in magnetotactic bacteria. FEMS Microbiology Reviews, 36(1): 232-255.
DOI:10.1111/j.1574-6976.2011.00315.x |
Kopp R E, Kirschvink J L. 2008. The identification and biogeochemical interpretation of fossil magnetotactic bacteria. Earth-Science Reviews, 86(1-4): 42-61.
DOI:10.1016/j.earscirev.2007.08.001 |
Koziaeva V V, Alekseeva L M, Uzun M M, Leão P, Sukhacheva M V, Patutina E O, Kolganova T V, Grouzdev D S. 2020. Biodiversity of magnetotactic bacteria in the freshwater lake Beloe Bordukovskoe, Russia. Microbiology, 89(3): 348-358.
DOI:10.1134/s002626172003008x |
Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33(7): 1870-1874.
DOI:10.1093/molbev/msw054 |
Lefèvre C T, Bazylinski D A. 2013. Ecology, diversity, and evolution of magnetotactic bacteria. Microbiology and Molecular Biology Reviews, 77(3): 497-526.
DOI:10.1128/mmbr.00021-13 |
Lefèvre C T, Bernadac A, Kui Y Z, Pradel N, Wu L F. 2009. Isolation and characterization of a magnetotactic bacterial culture from the Mediterranean Sea. Environmental Microbiology, 11(7): 1646-1657.
DOI:10.1111/j.1462-2920.2009.01887.x |
Lefèvre C T, Menguy N, Abreu F, Lins U, Pósfai M, Prozorov T, Pignol D, Frankel R B, Bazylinski D A. 2011. A cultured greigite-producing magnetotactic bacterium in a novel group of sulfate-reducing bacteria. Science, 334(6063): 1720-1723.
DOI:10.1126/science.1212596 |
Lefèvre C T, Viloria N, Schmidt M L, Pósfai M, Frankel R B, Bazylinski D A. 2012. Novel magnetite-producing magnetotactic bacteria belonging to the Gammaproteobacteria. The ISME Journal, 6(2): 440-450.
DOI:10.1038/ismej.2011.97 |
Lefèvre C T, Wu L F. 2013. Evolution of the bacterial organelle responsible for magnetotaxis. Trends in Microbiology, 21(10): 534-543.
DOI:10.1016/j.tim.2013.07.005 |
Li J H, Benzerara K, Bernard S, Beyssac O. 2013. The link between biomineralization and fossilization of bacteria: insights from field and experimental studies. Chemical Geology, 359: 49-69.
DOI:10.1016/j.chemgeo.2013.09.013 |
Li J H, Ge X, Zhang X K, Chen G J, Pan Y X. 2010. Recover vigorous cells of Magnetospirillum magneticum AMB-1 by capillary magnetic separation. Chinese Journal of Oceanology and Limnology, 28(4): 826-831.
DOI:10.1007/s00343-010-9068-4 |
Li J H, Liu P Y, Wang J, Roberts A P, Pan Y X. 2020a. Magnetotaxis as an adaptation to enable bacterial shuttling of microbial sulfur and sulfur cycling across aquatic oxicanoxic interfaces. Journal of Geophysical Research: Biogeosciences, 125(12): e2020JG006012.
DOI:10.1029/2020JG006012 |
Li J H, Menguy N, Arrio M A, Sainctavit P, Juhin A, Wang Y Z, Chen H T, Bunau O, Otero E, Ohresser P, Pan Y X. 2016. Controlled cobalt doping in the spinel structure of magnetosome magnetite: new evidences from elementand site-specific X-ray magnetic circular dichroism analyses. Journal of The Royal Society Interface, 13(121): 20160355.
DOI:10.1098/rsif.2016.0355 |
Li J H, Menguy N, Roberts A P, Gu L, Leroy E, Bourgon J, Yang X A, Zhao X, Liu P Y, Changela H G, Pan Y X. 2020b. Bullet-shaped magnetite biomineralization within a magnetotactic deltaproteobacterium: implications for magnetofossil identification. Journal of Geophysical Research: Biogeosciences, 125(7): e2020JG005680.
DOI:10.1029/2020jg005680 |
Li J H, Pan Y X. 2012. Environmental factors affect magnetite magnetosome synthesis in Magnetospirillum magneticum AMB-1: Implications for biologically controlled mineralization. Geomicrobiology Journal, 29(4): 362-373.
DOI:10.1080/01490451.2011.565401 |
Li J H, Zhang H, Liu P Y, Menguy N, Roberts A P, Chen H T, Wang Y Z, Pan Y X. 2019. Phylogenetic and structural identification of a novel magnetotacticDeltaproteobacteria strain, WYHR-1, from a freshwater lake. Applied and Environmental Microbiology, 85(14): e00731-19.
DOI:10.1128/aem.00731-19 |
Li J H, Zhang H, Menguy N, Benzerara K, Wang F X, Lin X T, Chen Z B, Pan Y X. 2017. Single-cell resolution of uncultured magnetotactic bacteria via fluorescencecoupled electron microscopy. Applied and Environmental Microbiology, 83(12): e00409-17.
DOI:10.1128/aem.00409-17 |
Lin W, Bazylinski D A, Xiao T, Wu L F, Pan Y X. 2014. Life with compass: diversity and biogeography of magnetotactic bacteria. Environmental Microbiology, 16(9): 2646-2658.
DOI:10.1111/1462-2920.12313 |
Lin W, Li J H, Schüler D, Jogler C, Pan Y X. 2009. Diversity analysis of magnetotactic bacteria in Lake Miyun, northern China, by restriction fragment length polymorphism. Systematic and Applied Microbiology, 32(5): 342-350.
DOI:10.1016/j.syapm.2008.10.005 |
Lin W, Pan Y X, Bazylinski D A. 2017. Diversity and ecology of and biomineralization by magnetotactic bacteria. Environmental Microbiology Reports, 9(4): 345-356.
DOI:10.1111/1758-2229.12550 |
Lin W, Pan Y X. 2015. A putative greigite-type magnetosome gene cluster from the candidate phylum Latescibacteria. Environmental Microbiology Reports, 7(2): 237-242.
DOI:10.1111/1758-2229.12234 |
Lin W, Wang Y Z, Li B, Pan Y X. 2012. A biogeographic distribution of magnetotactic bacteria influenced by salinity. The ISME Journal, 6(2): 475-479.
DOI:10.1038/ismej.2011.112 |
Lin W, Zhang W S, Zhao X, Roberts A P, Paterson G A, Bazylinski D A, Pan Y X. 2018. Genomic expansion of magnetotactic bacteria reveals an early common origin of magnetotaxis with lineage-specific evolution. The ISME Journal, 12(6): 1508-1519.
DOI:10.1038/s41396-018-0098-9 |
Lins U, Freitas F, Keim C N, Farina M. 2000. Electron spectroscopic imaging of magnetotactic bacteria: magnetosome morphology and diversity. Microscopy and Microanalysis, 6(5): 463-470.
DOI:10.1007/S100050010047 |
Lins U, Keim C N, Evans F F, Farina M, Buseck P R. 2007. Magnetite (Fe3O4) and greigite (Fe3S4) crystals in multicellular magnetotactic prokaryotes. Geomicrobiology Journal, 24(1): 43-50.
DOI:10.1080/01490450601134317 |
Liu J, Zhang W Y, Li X G, Li X G, Chen X M, Li J H, Teng Z J, Xu C, Santini C L, Zhao L, Zhao Y, Zhang H, Zhang W J, Xu K D, Li C L, Pan Y X, Xiao T, Pan H M, Wu L F. 2017. Bacterial community structure and novel species of magnetotactic bacteria in sediments from a seamount in the Mariana volcanic arc. Scientific Reports, 7(1): 17964.
DOI:10.1038/s41598-017-17445-4 |
Liu P Y, Liu Y, Zhao X, Roberts A P, Zhang H, Zheng Y, Wang F X, Wang L S, Menguy N, Pan Y X, Li J H. 2021. Diverse phylogeny and morphology of magnetite biomineralized by magnetotactic cocci. Environmental Microbiology, 23(2): 1115-1129.
DOI:10.1111/1462-2920.15254 |
Maloof A C, Kopp R E, Grotzinger J P, Fike D A, Bosak T, Vali H, Poussart P M, Weiss B P, Kirschvink J L. 2007. Sedimentary iron cycling and the origin and preservation of magnetization in platform carbonate muds, Andros Island, Bahamas. Earth and Planetary Science Letters, 259(3-4): 581-598.
DOI:10.1016/j.epsl.2007.05.021 |
Martins J L, Silveira T S, Silva K T, Lins U. 2009. Salinity dependence of the distribution of multicellular magnetotactic prokaryotes in a hypersaline lagoon. International Microbiology, 12(3): 193-201.
|
Nakamura C, Sakaguchi T, Kudo S, Burgess J G, Sode K, Matsunaga T. 1993. Characterization of iron uptake in the magnetic bacterium Aquaspirillum sp.AMB-1. Applied Biochemistry and Biotechnology, 39(1): 169-176.
DOI:10.1007/BF02918987 |
Neef A. 1997. Anwendung der in situ-Einzelzell-Identifizierung von Bakterien zur Populations-analyse in komplexen mikrobiellen Biozönosen. Technical University Munich, Munich, Germany.
|
Pan H M, Zhu K L, Song T, Kui Y Z, Lefèvre C, Xing S E, Liu M, Zhao S J, Xiao T, Wu L F. 2008. Characterization of a homogeneous taxonomic group of marine magnetotactic cocci within a low tide zone in the China Sea. Environmental Microbiology, 10(5): 1158-1164.
DOI:10.1111/j.1462-2920.2007.01532.x |
Pan Y X, Petersen N, Davila A F, Zhang L M, Winklhofer M, Liu Q S, Hanzlik M, Zhu R X. 2005. The detection of bacterial magnetite in recent sediments of Lake Chiemsee (southern Germany). Earth and Planetary Science Letters, 232(1-2): 109-123.
DOI:10.1016/j.epsl.2005.01.006 |
Pernthaler A, Pernthaler J, Amann R. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Applied and Environmental Microbiology, 68(6): 3094-3101.
DOI:10.1128/AEM.68.6.3094-3101.2002 |
Petersen N, von Dobeneck T, Vali H. 1986. Fossil bacterial magnetite in deep-sea sediments from the South Atlantic Ocean. Nature, 320(6063): 611-615.
DOI:10.1038/320611a0 |
Pósfai M, Lefèvre C T, Trubitsyn D, Bazylinski D A, Frankel R B. 2013. Phylogenetic significance of composition and crystal morphology of magnetosome minerals. Frontiers in Microbiology, 4: 344.
DOI:10.3389/fmicb.2013.00344 |
Qian X X, Santini C L, Kosta A, Menguy N, Guenno H L, Zhang W Y, Li J H, Chen Y R, Liu J, Alberto F, Espinosa L, Xiao T, Wu L F. 2020. Juxtaposed membranes underpin cellular adhesion and display unilateral cell division of multicellular magnetotactic prokaryotes. Environmental Microbiology, 22(4): 1481-1494.
DOI:10.1111/1462-2920.14710 |
Rivas-Lamelo S, Benzerara K, Lefèvre C T, Monteil C L, Jézéquel D, Menguy N, Viollier E, Guyot F, Férard C, Poinsot M, Skouri-Panet F, Trcera N, Miot J, Duprat E. 2017. Magnetotactic bacteria as a new model for P sequestration in the ferruginous Lake Pavin. Geochemical Perspectives Letters, 5: 35-41.
DOI:10.7185/geochemlet.1743 |
Schübbe S, Williams T J, Xie G, Kiss H E, Brettin T S, Martinez D, Ross C A, Schü ler D, Cox B L, Nealson K H, Bazylinski D A. 2009. Complete genome sequence of the chemolithoautotrophic marine Magnetotactic Coccus Strain MC-1. Applied Environmental Microbiology, 75(14): 4835-4852.
DOI:10.1128/aem.02874-08 |
Schüler D, Köhler M. 1992. The isolation of a new magnetic spirillum. Zentralblatt für Mikrobiologie, 147(1-2): 150-151.
DOI:10.1016/S0232-4393(11)80377-X |
Schulz-Vogt H N, Pollehne F, Jürgens K, Arz H W, Beier S, Bahlo R, Dellwig O, Henkel J V, Herlemann D P R, Krüger S, Leipe T, Schott T. 2019. Effect of large magnetotactic bacteria with polyphosphate inclusions on the phosphate profile of the suboxic zone in the Black Sea. The ISME Journal, 13(5): 1198-1208.
DOI:10.1038/s41396-018-0315-6 |
Simmons S L, Edwards K J. 2007. Unexpected diversity in populations of the many-celled magnetotactic prokaryote. Environmental Microbiology, 9(1): 206-215.
DOI:10.1111/j.1462-2920.2006.01129.x |
Spring S, Amann R, Ludwig W, Schleifer K H, Van Gemerden H, Petersen N. 1993. Dominating role of an unusual magnetotactic bacterium in the microaerobic zone of a freshwater sediment. Applied and Environmental Microbiology, 59(8): 2397-2403.
DOI:10.1128/AEM.59.8.2397-2403.1993 |
Taoka A, Kondo J, Oestreicher Z, Fukumori Y. 2014. Characterization of uncultured giant rod-shaped magnetotactic Gammaproteobacteria from a freshwater pond in Kanazawa, Japan. Microbiology, 160(10): 2226-2234.
DOI:10.1099/mic.0.078717-0 |
Wang Y Z, Lin W, Li J H, Pan Y X. 2012. High diversity of magnetotactic Deltaproteobacteria in a freshwater niche. Applied and Environmental Microbiology, 79(8): 2813-2827.
DOI:10.1128/AEM.03635-12 |
Wang Y Z, Lin W, Li J H, Pan Y X. 2013. Changes of cell growth and magnetosome biomineralization in Magnetospirillum magneticum AMB-1 after ultraviolet-B irradiation. Frontiers in Microbiology, 4: 397.
DOI:10.3389/fmicb.2013.00397 |
Wenter R, Wanner G, Schüler D, Overmann J. 2009. Ultrastructure, tactic behaviour and potential for sulfate reduction of a novel multicellular magnetotactic prokaryote from North Sea sediments. Environmental Microbiology, 11(6): 1493-1505.
DOI:10.1111/j.1462-2920.2009.01877.x |
Wolfe R S, Thauer R K, Pfennig N. 1987. A 'capillary racetrack' method for isolation of magnetotactic bacteria. FEMS Microbiology Ecology, 3(1): 31-35.
DOI:10.1111/j.1574-6968.1987.tb02335.x |
Zhang H, Menguy N, Wang F X, Benzerara K, Leroy E, Liu P Y, Liu W Q, Wang C L, Pan Y X, Chen Z B, Li J H. 2017. Magnetotactic coccus strain SHHC-1 affiliated to Alphaproteobacteria forms octahedral magnetite magnetosomes. Frontiers in Microbiology, 8: 969.
DOI:10.3389/fmicb.2017.00969 |
Zhang R, Chen Y R, Du H J, Zhang W Y, Pan H M, Xiao T, Wu L F. 2014. Characterization and phylogenetic identification of a species of spherical multicellular magnetotactic prokaryotes that produces both magnetite and greigite crystals. Research in Microbiology, 165(7): 481-489.
DOI:10.1016/j.resmic.2014.07.012 |
Zhou K, Pan H M, Zhang S D, Yue H D, Xiao T, Wu L F. 2011. Occurrence and microscopic analyses of multicellular magnetotactic prokaryotes from coastal sediments in the Yellow Sea. Chinese Journal of Oceanology and Limnology, 29(2): 246-251.
DOI:10.1007/s00343-011-0032-8 |
Zhou K, Zhang W Y, Pan H M, Li J H, Yue H D, Xiao T, Wu L F. 2013. Adaptation of spherical multicellular magnetotactic prokaryotes to the geochemically variable habitat of an intertidal zone. Environmental Microbiology, 15(5): 1595-1605.
DOI:10.1111/1462-2920.12057 |
Zhu K L, Pan H M, Li J H, Kui Y Z, Zhang S D, Zhang W Y, Zhou K, Yue H D, Pan Y X, Xiao T, Wu L F. 2010. Isolation and characterization of a marine magnetotactic spirillum axenic culture QH-2 from an intertidal zone of the China Sea. Research in Microbiology, 161(4): 276-283.
DOI:10.1016/j.resmic.2010.02.003 |
 2021, Vol. 39
2021, Vol. 39