Institute of Oceanology, Chinese Academy of Sciences
Article Information
- LIU Yang, CHEN Youxin, FANG Haiyan, LU Hanyang, WU Xingqiang, YU Gongliang, NAKANO Shin-ichi, LI Renhui
- Relationship between morphospecies and microcystinproducing genotypes of Microcystis species in Chinese freshwaters
- Journal of Oceanology and Limnology, 39(5): 1926-1937
- http://dx.doi.org/10.1007/s00343-020-0276-2
Article History
- Received Jul. 22, 2020
- accepted in principle Sep. 29, 2020
- accepted for publication Nov. 10, 2020
2 Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan 430072, China;
3 Center for Ecological Research, Kyoto University, Shiga 520-2113, Japan;
4 College of Life and Environmental Sciences, Wenzhou University, Wenzhou 325000, China;
5 Journal of Henan Normal University, Xinxiang 453007, China
Cyanobacterial blooms are frequently observed in eutrophic or estuarine waters worldwide. Several genera of cyanobacteria, including Microcystis, Anabaena, Planktothrix, Aphanizomenon, and Nostoc, can produce a variety of secondary metabolites such as hepatotoxins, neurotoxins, and cytotoxins (Liu et al., 2016; Shan et al., 2019; Wang et al., 2019, 2021). Among cyanobacteria, Microcystis is considered the most prominent contributor to the production of microcystin (MC), which are potent hepatotoxins. More than 90 MC variants have been reported so far (Hotto et al., 2008; Attard et al., 2018). These toxins could potentially inhibit the activities of eukaryotic serine/threonine protein phosphatase 1 and 2A, and have been implicated in several livestock and human poisonings (Mikalsen et al., 2003; Tanabe et al., 2004). Thus, the World Health Organization has published a preliminary guideline value for microcystinLR (MC-LR) in drinking water, which is 1 μg/L (WHO, 1998; Wu et al., 2007; Otten and Paerl, 2011).
MCs are produced by a mixed polyketide synthase (PKS)/nonribosomal peptide synthetase (NRPS) complex via a thiotemplate mechanism (Mikalsen et al., 2003). MC synthetase (mcy) gene clusters encoding these biosynthetic enzymes have been identified and sequenced from the unicellular organism Microcystis aeruginosa (Rantala et al., 2004). The mcy gene cluster is composed of 10 genes arranged bidirectionally in two operons (mcyA-C and mcyD-J) (Rantala et al., 2004; Cirés et al., 2013). The mcyA-J gene cluster is essential for the production of MCs by Microcystis. Therefore, molecular probes have been developed to detect and study the community composition and dynamics of MCproducing and non-MC-producing Microcystis strains in the field (Kurmayer et al., 2002; Via-Ordorika et al., 2004). The mcyB gene, which encodes a peptide synthetase of 242 kDa, has been widely used as a marker for toxin biosynthesis and genetic diversity studies (Mikalsen et al., 2003; Dyble, 2008). In contrast, a small number of Microcystis strains tested positive for the mcy gene but lacked detectable MCs. Insertional inactivation, homology of the mcyB region or even the presence of the mcy gene cluster in some nontoxic Microcystis strains may explain these discrepancies (Via-Ordorika et al., 2004). Therefore, molecular methods combining chemical detection are necessary. These methods have been widely used to detect MC-producing Microcystis populations.
Likewise, these methods are helpful for the rapid detection of the toxic Microcystis in freshwater bodies, and also provide a support for the studies on the relationships among the morphology, colony size, and the occurrence of mcy gene-containing colonies. Relationship between Microcystis colony size and the occurrence of mcy gene-containing colonies was studied earlier (Via-Ordorika et al., 2004; Dyble et al., 2008). By far, a consensus is that the largest size colony has a maximum proportion of mcy (Jungmann et al., 1996; Via-Ordorika et al., 2004). A new study of the occurrence of MCs biosynthesis genes in natural populations of different Microcystis species in Russian reservoirs (Sidelev et al., 2020) show that no correlation between colony size and the frequency of mcy genes for individual morphospecies M. aeruginosa and M. flos-aquae. However, they identified only five morphotypes of Microcystis and two of them were chosen in 10 reservoirs. At present, at least 51 species of Microcystis have been reported in the world, and approximately 13 species have been described in China (Otsuka et al., 1999a; Wu et al., 2007; Xu et al., 2008; Xiao et al., 2017; Pérez-Carrascal et al., 2019). Therefore, more morphospecies and water bodies are needed to be used to investigate and analyze the morphology and toxigenic capacity of Microcystis species. At present, Microcystis are dominant species in most freshwater bodies of China, such as Taihu Lake, Caohu Lake, and Dianchi Lake (Xu et al., 2008; Shen et al., 2018). Most species of the genus Microcystis have the ability to produce MCs, and a quick judgment to identify potential toxic Microcystis blooms by morphology is also needed for water quality management. In this study, 20 water bodies (in 13 cities) across several climatic zones of China were surveyed to examine the relationship between morphospecies and MC-producing genotypes of the genus Microcystis. Furthermore, the mcy genes and toxin content were also analyzed by Polymerase Chain Reactio (PCR) and high-performance liquid chromatography (HPLC).
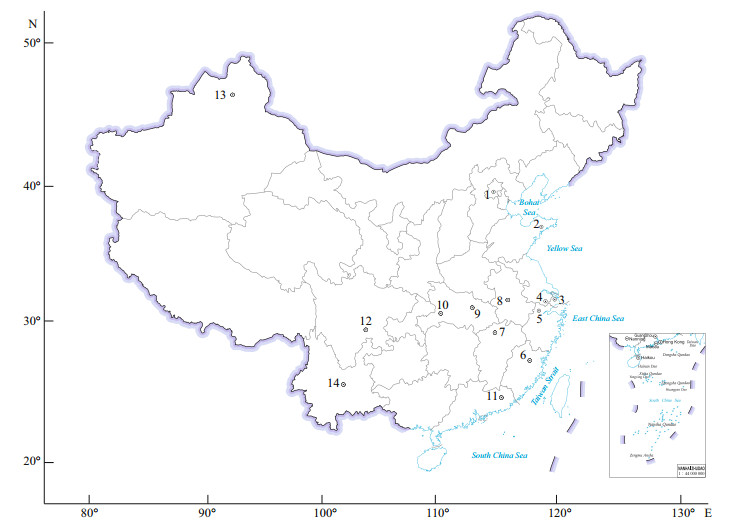
2 MATERIAL AND METHOD 2.1 Sampling and isolationSamples were collected with a plankton net (25#, 200 mesh) from 20 freshwater bodies, located in 13 cities of China (Fig. 1) from tropical to temperate zones during 2006 and 2007. There are two types of samples. One is from field experiments and the other is from laboratory experiments. In the field experiment, a 500-mL sample was collected from each water body, and all the types of Microcystis from each water body were distinguished by classical taxonomic identification. Fifty samples of each Microcystis type were selected for whole-cell detection. In the laboratory experiment, single colonies of Microcystis from each water body were isolated using the Pasteur capillary pipette method under a dissecting microscope, and each colony that was chosen was washed three times in sterile water, and each colony was kept in one well of a 24-well plate in 2-mL MA culture medium (Ichimura, 1979). To successfully isolate single colonies of Microcysits, we picked up 24 colonies in each freshwater body based on our experience. All cultures were grown at (25±1) ℃ with a 12-h꞉12-h light꞉dark cycle under a photon irradiance of 25 μmol photons/(m2·s) provided by daylight fluorescent lamps. Taxonomic identifications were assigned following the descriptions of Komárek and Anagnostidis (1998) by a light microscope (Olympus BX51, Japan).
|
| Fig.1 Map of the sampling sites The number represents different sampling sites, including 20 water bodies. 1: Beihai Lake, Beijing; 2: Jihongtan Reservoir, Qingdao, Shandong; 3: Dishui Lake, Shanghai; 4: Taihu Lake, Jiangshu; 5: Fuchun River Hangzhou, and Guazhu Pond, Shaoxing, Zhejiang; 6: Baima River, Fuzhou, Fujian; 7: Majianggong River, Jiangxi; 8: Chaohu Lake, Anhui; 9: Donghu Lake, Nanhu Lake, Chidong Lake, Guanqiao Pond, Wuhan, Hubei; 10: the Three Gorges, Yichang, Hubei; 11: Zhongshan Lake, Guangdong; 12: the Three Gorges, Sichuan; 13: Wulungu Lake, Xinjiang; 14: Fuxian Lake, Xingyun Lake, Dianchi Lake, Yunnan. Map review No. GS(2019)1682. |
Genomic DNAs (the pure culture grew to a certain biomass) were extracted following the modified cetyltrimethylammonium bromide (CTAB) method as described by Meißner et al. (1996). The primers for amplification of the Microcystis 16S rDNA fragment were MicroF (5′-GCCGCRAGGT-GAAAMCTAA-3′) and MicroR (5′-AATCCAAAG-ACCTTCCTCCC-3′) (Neilan et al., 1997). To identify the micorcystinproducing ability of Microcystis strains, we detected a region within the mcyB gene required for microcystin biosynthesis by PCR. The primer sequences were tox4f (5′-GGATATCCTC-TCAGATTCGG-3′) and tox4r (5′-CACTAACCC-CTATTTTGGATACC-3′) (Kurmayer et al., 2002), which have been suggested to be specific for the genus Microcystis. The PCRs were carried out in 50-μL mixtures containing 5–10 ng of genomic DNA, 25 μL of 2×PCR Master Mix, 10 pmol of primer and sterile water to 50 μL. The reactions were run in a BioRad Thermal Cycler (BioRad, USA) with one cycle at 94 ℃ for 5 min, 35 cycles at 94 ℃ for 30 s, annealing at 50 ℃ for 30 s (MicroF/MicroR) or at 55 ℃ for 30 s (tox4f/tox4r), strand extension at 72 ℃ for 25 s, and final extension at 72 ℃ for 5 min. PCR products were detected by 1% agarose gel electrophoresis, and amplified bands were detected under 300 nm ultraviolet light. Then, PCR products were purified with the kits (Qiagen, Germany) according to the manufacturer's protocol. Then, the purified products were cloned into the PMD18-T vector (TaKaRa, Japan) and transformed into E. coli DH5α competent cells (TaKaRa, Japan) on an ice bath. Finally, a single positive clone was selected for sequencing (Invitrogen, Shanghai, China) for each isolate. The accession numbers for the obtained mcyB sequences are KJ818165–KJ818201, and for 16S rDNA gene sequences are KJ818122- KJ818163.
2.3 Sequence alignment and phylogenetic analysisSequences were initially corrected and aligned by using BioEdit software (http://www.mbio.ncsu.edu/bioedit/bioedit.html). Twenty-one strains of Microcystis covering 6 different species were downloaded from GenBank (www.blast.ncbi.nlm. nih.gov), and 37 sequences obtained from this study were used for the 16S rDNA phylogenetic analysis. The 16S rDNA sequence of Synechocystis PCC6803 was used as the out-group. Phylogenetic distances in the trees were estimated using the algorithm of Kimura (1980), and 1 000 bootstrap trials were performed. The phylogenetic tree was constructed using the neighbor-joining (NJ) method in MEGA X (Kumar et al., 2018). Phylogenetic analysis for mcyB was performed the same as above. A total of 67 sequences were used to construct the phylogenetic tree, including 24 sequences from GenBank, and 42 sequences from the present study, and Nostoc sp. was used as the outgroup. Finally, the intra-group and between group similarity were performed by using "sequence identity matrix" option in BioEdit software.
2.4 HPLC analysisStrains of Microcystis that have been shown to have the mcyB fragment were large-scale cultured and collected, as previously described by Liu et al. (2011). The Microcystis cells were lyophilized, and approximately 50 mg of these freeze-dried cells were extracted for the analysis of MCs. Chromatographic separation was performed using a Waters Alliance 2695 HPLC system, Waters 2996 photo diode array detector and Waters empower chromatography software (Waters, USA). The wavelength was 238 nm, and the chromatographic column was a Synergi Hydro-RP C18 column (4 μm, 250 mm×4.6 mm). The mobile phase consisted of pure methanol (solvent A) and 0.05% aqueous trifluoroacetic acid (solvent B). The linear gradient was 50% solvent A at 0 min, 80% solvent A at 15 min, 50% solvent A at 16 min, and 50% solvent A at 20 min. The detector resolution was set at 1.2 nm and the flow rate was set at 1 mL/min with a column temperature of 30 ℃.
2.5 Whole cell PCR in field samplesA whole cell PCR method was performed to increase the efficiency of detecting MC from field samples (Howitt, 1996; Pan et al., 2001). Microcysits colony for the whole-cell PCR was isolated from each sample by using the Pasteur capillary pipette method. The isolates were washed one to three times with distilled water and rechecked under an inverted microscope. Finally, 1–2-μL isolates as the DNA template were directly used in PCR amplification to detect the mcyB gene. The method used was the same as that mentioned in Section 2.2. Finally, 50 samples were collected and detected to evaluate the ratio of MC producing strains in the field.
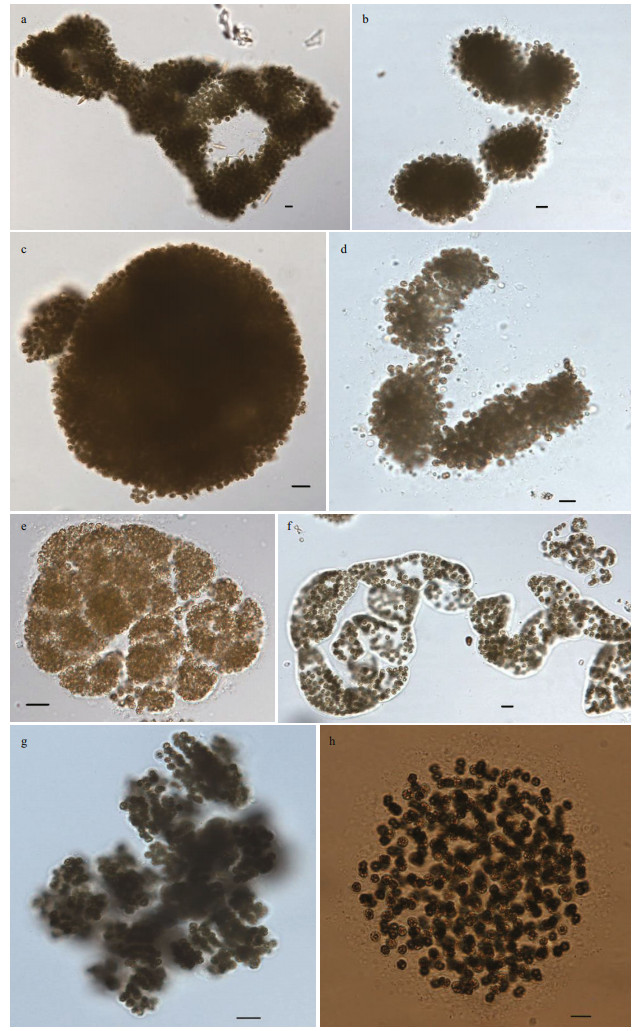
3 RESULT 3.1 Morphospecies of 186 Microcystis strainsA total of 186 strains of Microcystis were isolated from 20 water bodies in China. The strains were distinguished by classical taxonomic identification, since it is still an irreplaceable method. It is necessary to identify the strains as soon as they were collected due to their high phenotypic plasticity. Finally, eight morphospecies were identified according to the descriptions of Komárek and Komárková (2002) (Fig. 2): M. aeruginosa (Kuëtzing) Kuëtzing, M. botrys Teiling, M. flos-aquae (Wittrock) Kirchner, M. novacekii (Komaèrek) Compère, M. ichthyoblabe Kuëtzing, M. wesenbergii (Komárek) Komárek, M. viridis (A. Brown) Lemmermann, and M. smithii Komárek & Anagnostidis. The number of each morphospecies was shown in Table 1.
|
| Fig.2 Colonies of eight Microcystis morphospecies in China a. M. aeruginosa; b. M. botrys; c. M. flos-aquae; d. M. novacekii; e. M. ichthyoblabe; f. M. wesenbergii; g. M. viridis; h. M. smithii. Scale bars=10 μm. |
|
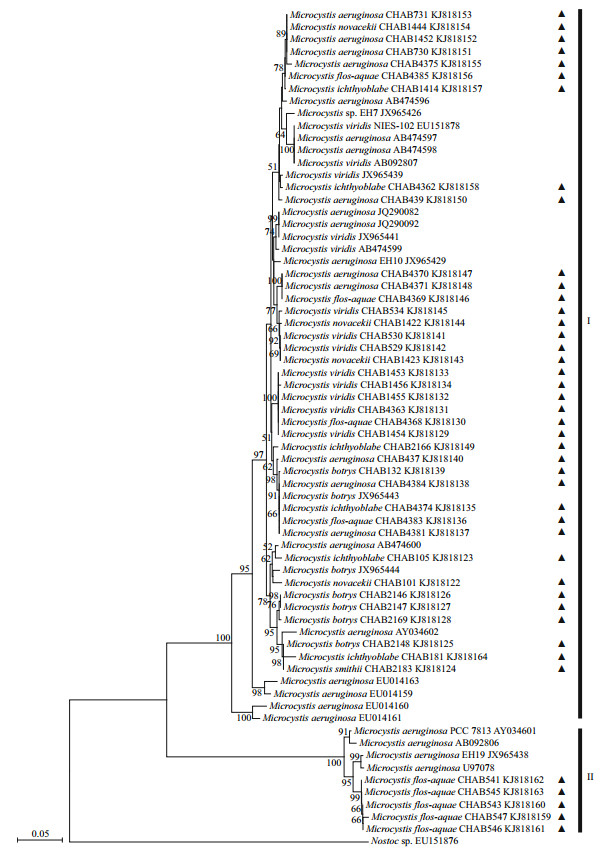
From the 186 isolates, only 43 showed the presence of mcyB by PCR. The 16S rRNA gene sequences of the 43 Microcystis strains with mcyB genes were amplified. Each sequence showed a product of 1 444 bp, which corresponded to the length of the 16S rDNA, and an alignment of 1 402 nucleotides after excluding positions with gaps was used. The NJ phylogenetic tree based on the 16S rRNA gene (Fig. 3), includes all 43 strains of Microcystis in this study and 21 strains of Microcystis obtained from GenBank. Amplification of the mcyB gene with primers tox4f and tox4r revealed that 43 strains of Microcystis gave PCR products in size of 1 312 bp and an alignment of 1 116 nucleotides after excluding positions with gaps was used. The NJ phylogenetic tree based on the mcyB gene (Fig. 4) includes all 43 strains of Microcystis in this study and 24 strains of Microcystis obtained from GenBank. Finally, the sequence similarities in Goup I and Group II (Fig. 4) were calculated separately. The high similarity within each group was 99%, and the maximum similarity of mcyB sequences between the two groups was 69.1%.

|
| Fig.3 Phylogenetic tree (NJ) based on 16S rRNA sequences Synechocystis sp. PCC6803 was used as the outgroup. The numbers at each node represent the percentage of bootstrap setting as 1 000, only showing those above 50%. The isolates obtained in this study are in bold fonts. |
|
| Fig.4 Phylogenetic tree (NJ) based on mcyB sequences Nostoc sp. was used as the outgroup. The numbers at each node represent the percentage of bootstrap setting as 1 000, only showing those above 50%. Triangle represents the strains obtained in this study. |
Analysis of the mcyB gene from 186 strains of Microcystis by PCR showed that only 43 strains produced a PCR product for mcyB. Then, strains of the same region and morphology were removed (strains from the same place and sampling site were grouped, and 1–2 isolates were randomly selected), and 20 strains were finally enlarged in large-scale culture to analyze the type and content of microcystin in different morphospecies. The results (Table 1) show that 7 morphospecies sampled from 10 different waterbodies contained different microcystin variants and contents. All tested M. viridis contained two types of microcystin (MC-LR and MC-RR) except the strain from Fuxian Lake. The contents of MC-RR were much higher in M. viridis than MC-LR. All M. flos-aquae strains contained MC-LR but no MC-RR was observed.
3.4 The proportion of Microcystis containing the mcyB geneIndividual Microcystis colonies, consisting of 8 different morphospecies from indoor culture and field samples, were tested for the mcyB gene using a wholecell PCR assay. Thirty individual Microcystis colonies of each morphospecies were used to detect the mcyB gene. The results are shown in Table 2. All of the M. wesenbergii morphospecies produced no MC, which is consistent with previous studies (Xu et al., 2008). In our survey, the proportion of four morphospecies (M. viridis, M. ichthyoblabe, M. novacekii, and M. aeruginosa) that contained the mcyB gene exceeded 50% in the field samples. Accordingly, these species are most likely to produce MCs in Chinese water bodies.
Although cyanobacterial blooms have broken out all over the world, the compositions of Microcystis in different regions were varied. Six morphospecies of Microcystis were distinguished in Japanese waters (Otsuka et al., 1999b), five morphotypes of Microcystis were identified in Russia (Sidelev et al., 2020), and seven morphospecies were found in European waters (Kurmayer et al., 2002; Via-Ordorika et al., 2004). The eight species identified in this study were strains common in most water bodies in China and could produce MCs, except for M. wesenbergii. This result is consistent with previous studies in that no mcyB fragment was detected (Xu et al., 2008). Although the seven species were able to produce MCs, some of them may lack detectable MCs (Tanabe et al., 2004; Via-Ordorika et al., 2004). It was still unclear to what extend the morphological characteristics were linked to the ability of producing MCs. However, the colonies assigned to M. aeruginosa were identified as having the highest frequency of toxin production (Kurmayer et al., 2002; Via-Ordorika et al., 2004). A previous research found that toxicity of Microcystis under natural conditions could be associated mainly with large-diameter colonies (Jungmann et al., 1996), which may be consistent with our finding that M. aeruginosa always had a large size in the field.
4.2 Phylogenetic analysisThe phylogenetic tree (Fig. 3) shows low bootstrap values as a result of the extremely high similarity (99.2%–100%) of sequences determined. The species of Microcystis isolated from different places were mixed together according to the topology tree. Moreover, different morphotype strains were also mixed; for example, M. botrys, M. viridis, and M. wesenbergii were mixed with the "M. aeruginosa complex" (Otsuka et al., 1998, 1999a; Tan et al., 2010). Although the 16S rDNA sequence is insufficiently variable for phylogenetic analysis of Microcystis spp. at the species level, it is still valid at the genus level. Therefore, this may appeal to the findings of population genomics that predicated Microcystis represents a single, globally distributed, and homogeneous gene pool (Pérez-Carrascal et al., 2019). However, our study agreed with the view that more "ecophenic" and/or phenotypic forms have been described than genospecies exist (Palinska et al., 1996; Le Ai Nguyen et al., 2012).
The phylogenetic tree (Fig. 4) could be divided into two groups (Groups I & II), and high similarity (99%) of sequences within each group was found. The maximum similarity of mcyB sequences between the two groups was 69.1%. Group I covered more of the sequences in GenBank than Group II. A large proportion of species in Group II were uncultured cyanobacteria. The three strains of M. flos-aquae in Group II (CHAB543, CHAB546, and CHAB547) contained one type of MC-LR but no MC-RR. The mcyB region was more variable in the MC biosynthesis gene cluster and showed high mutation rates (Kurmayer et al., 2002; Mikalsen et al., 2003). This was related to the production of various MC isoforms. We use the same sequences from GenBank (AY034602 and AB092806) that covered the MC synthetase module encoding the mcyB1 domain to perform phylogenetic analysis (Dyble et al. 2008). The results of this study confirm that mcyB sequences in Microcystis could be grouped into two major clusters (mcyB1(B) and mcyB1(C)) (Mikalsen et al., 2003). The (B) variants of mcyB1 produced various MC-LR isoforms. The (C) variant of the mcyB1 module synthesized MC-RR, and the members of some subgroups produced MC-LR in addition to MC-RR (Mikalsen et al., 2003; Dyble et al., 2008; Otten and Paerl, 2011). These findings were very interesting as Group I and Group II have mcyB1(C) and mcyB1(B), respectively, each group with the same properties to produce MC isoforms. Such findings suggested that the genotypic variations within the mcyB module were strongly correlated with their isoforms of MC.
4.3 Analysis of the mcyB gene and MCsThe mcyB sequences of these M. flos-aquae were also unique and had a low similarity with others in this study. Previous studies have revealed several phylogenetic subgroups of the adenylation domain in the mcyB gene. Mikalsen et al. (2003) named the two major clusters of mcyB sequences in Microcystis mcyB1(B) and mcyB1(C). Strains that synthesized MC-RR generally belonged to the C-type subgroup, and the B-type subgroup generally produced MC-LR. This was the reason why these strains were divided into two groups in the phylogenetic tree. Some strains from GenBank could group with these M. flos-aquae strains, and whether they contain no MC-RR was unknown. The contents of MC-RR were much higher than that of MC-LR in Group I. These findings also revealed that geographical segregation existed on the basis of the homology of mcyB sequences. These conclusions generally supported the results of previous studies showing that Microcystis strains may have geographical distribution in the world (Wu et al., 2007). In our study, all strains of M. aeruginosa had both MC-LR and MC-RR. The M. botrys from different sites contained distinct MCs. It was also found that even Microcystis from the same place contained distinct MCs. Again, this study supported the results of previous studies showing that genetic variation in the mcyB1 module generated different MC isoforms (Mikalsen et al., 2003). This is the first to report two strains of M. novacekii producing both MC-LR and MC-RR that were isolated in China. Previous studies found that no M. novacekii strains isolated in Japan produced MC, but two toxic strains of M. novacekii were isolated in Thailand (Otsuka et al., 1999a, 2000; Tsujimura et al., 2000). Likewise, we found that M. smithii produced MC-RR, which we covered in previous article (Liu et al., 2011). Although the M. ichthyoblabe morphotype in Taihu Lake consistently lacked the mcyB gene, all four strains possessed the mcyB gene and produced both MC-LR and MC-RR except one strain. This strain (CHAB181) contained only MC-RR, but its MC content was the highest, up to 2 825.44 μg/g. Previously studies reported little about the characteristics of toxin production by M. ichthyoblabe, and the main reason may be that it was difficult to distinguish these four close morphospecies (M. aeruginosa, M. flos-aquae, M. ichthyoblabe, and M. novacekii) (Otsuka et al., 1999b; Tan et al., 2010). These four species were presumed to be "M. aeruginosa complex". M. aeruginosa has been extensively studied. All the strains in Table 1 are consistent with B-type variants of mcyB1 producing various MC-LR isoforms and most C-type variants of the mcyB1 module synthesizing MC-RR comparing with previous studies (Mikalsen et al., 2003; Dyble et al., 2008).
4.4 The proportion of the mcyB gene in cultured and field Microcystis in Chinese freshwatersKurmayer et al. (2002) reported that in Lake Wannsee of Germany, the proportion of Microcystis colonies with the mcyB gene were as follows: 73% M. aeruginosa, 16% M. ichthyoblabe, 37% M. flosaquae and 45% M. botrys. It was interesting that the proportion of toxic M. botrys and M. aeruginosa was almost the same. Another study in European freshwater bodies also found that more than 75% of the M. botrys and M. aeruginosa contained the mcy genes, and approximately 20% of the M. ichthyoblabe and M. vividis gave a PCR product of the mcy genes (Via-Ordorika et al., 2004). Although the proportion of toxic M. ichthyoblabe and M. vividis was different between China and Europe, M. aeruginosa was the most likely morphotype to produce MC (Vasconcelos and Pereira, 2001; Cirés et al., 2013). In this study, the proportions of M. flos-aquae and M. smithii in the field were 20% and 11%, respectively. We also studied the proportion of cultured species, and the results from the indoor cultures are slightly different from those of the field samples. We inferred that it might be associated with the culture conditions because temperature, light, and nutrients had some effects on the production of MCs (Song et al., 1998; Wiedner et al., 2003). Moreover, most Microcystis morphotypes could maintain their colony characteristics for approximately one month (Otsuka et al., 2000). Accordingly, it seemed to be obscure to find distinction among these morphospecies. This may support the idea of population genomics that Microcystis populations are not a coherent species (Pérez-Carrascal et al., 2019). Inconsistencies between traditional and population genomic classifications exist and need a further study. The strains of Microcystis used in this study were not sterile. Therefore, this may be another reason why the two results are different. However, our findings still provide new insight into Microcystis hazard assessment and field monitoring.
5 CONCLUSIONThe results show a relationship between morphospecies and MC-producing genotypes of common Microcystis species in Chinese freshwaters. The data also show that mcyB gene-containing Microcystis colonies of China could be divided into two groups (Group I and Group II). Most Microcystis strains of Group I contained two types of MC isoforms, while Group II contained only MC-LR. The proportion of four morphospecies (M. vividis, M. ichthyoblabe, M. novacekii, and M. aeruginosa) that contained the mcyB gene exceeded 50% in the field samples. It was consistent with previous studies that M. aeruginosa was the most likely morphotype to produce MC in the world. It could help water quality managers for a quick judgment on potential toxic Microcystis blooms.
6 DATA AVAILABILITY STATEMENTThe data generated in this study are available from the corresponding author on reasonable request.
7 ACKNOWLEDGMENTWe are grateful to the lab members from Harmful Alga Group for assistances with samples collection and strains isolation. We are also thankful to Dr. Shen LIN, Li YANG, Zhongjie WANG, Mengling ZHU, and Prof. Zhongxing WU for your help in this research.
Attard T J, Carter M D, Fang M X, Johnson R C, Reid G E. 2018. Structural characterization and absolute quantification of microcystin peptides using collision-induced and ultraviolet photo-dissociation tandem mass spectrometry. Journal of the American Society for Mass Spectrometry, 29(9): 1 812-1 825.
DOI:10.1021/jasms.8b05898 |
Cirés S, Wörmer L, Carrasco D, Quesada A. 2013. Sedimentation patterns of toxin-producing Microcystis morphospecies in freshwater reservoirs. Toxins, 5(5): 939-957.
DOI:10.3390/toxins5050939 |
Dyble J, Fahnenstiel G L, Litaker R W, Millie D F, Tester P A. 2008. Microcystin concentrations and genetic diversity of Microcystis in the Lower Great Lakes. Environmental Toxicology, 23(4): 507-516.
DOI:10.1002/tox.20370 |
Hotto A M, Satchwell M F, Berry D L, Gobler C J, Boyer G L. 2008. Spatial and temporal diversity of microcystins and microcystin-producing genotypes in Oneida Lake, NY. Harmful Algae, 7(5): 671-681.
DOI:10.1016/j.hal.2008.02.001 |
Howitt C A. 1996. Amplification of DNA from whole cells of cyanobacteria using PCR. Biotechniques, 21(1): 32-34.
DOI:10.2144/96211bm05 |
Ichimura T. 1979. Media for blue-green algae. In: Nishizawa K, Chihara M eds. Methods in Algalogical Studies. Kyouritsu, Tokyo, p. 294–305.
|
Jungmann D, Ludwichowski K U, Faltin V, Benndorf J. 1996. A field study to investigate environmental factors that could effect microcystin synthesis of a Microcystis population in the Bautzen reservoir. Internationale Revue der Gesamten Hydrobiologie und Hydrographie, 81(4): 493-501.
DOI:10.1002/iroh.19960810402 |
Kimura M. 1980. Kimura's two-parameter model of Models of DNA Evolution. In: Felsenstein J ed. 2004. Inferring Phylogenies. Sunderland, Massachusetts: Sinauer Associates, Inc.
|
Komárek J, Anagnostidis K. 1998. Cyanoprokaryota 1. Teil: chroococcales. In: Ettl H, Gärtner G, Heynig H, Mollenhauer D eds. Süßwasserflora von Mitteleuropa, Bd 19/1. Gustav Fischer, Jena, p. 164–190.
|
Komárek J, Komárková J. 2002. Review of the European Microcystis morphospecies (Cyanoprokaryotes) from nature. Fottea, 2(1): 1-24.
|
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35(6): 1 547-1 549.
DOI:10.1093/molbev/msy096 |
Kurmayer R, Dittmann E, Fastner J, Chorus I. 2002. Diversity of microcystin genes within a population of the toxic cyanobacterium Microcystis spp. in Lake Wannsee (Berlin, Germany). Microbial Ecology, 43(1): 107-118.
DOI:10.1007/s00248-001-0039-3 |
Le Ai Nguyen V, Tanabe Y, Matsuura H, Kaya K, Watanabe M M. 2012. Morphological, biochemical and phylogenetic assessments of water-bloom-forming tropical morphospecies of Microcystis (Chroococcales, Cyanobacteria). Phycological Research, 60(3): 208-222.
DOI:10.1111/j.1440-1835.2012.00650.x |
Liu Y, Tan W H, Wu X Q, Wu Z X, Yu G L, Li R H. 2011. First report of microcystin production in Microcystis smithii Komárek and Anagnostidis (Cyanobacteria) from a water bloom in Eastern China. Journal of Environmental Sciences, 23(1): 102-107.
DOI:10.1016/S1001-0742(10)60379-8 |
Liu Y, Xu Y, Wang Z J, Xiao P, Yu G L, Wang G X, Li R H. 2016. Dominance and succession of Microcystis genotypes and morphotypes in Lake Taihu, a large and shallow freshwater lake in China. Environmental Pollution, 219: 399-408.
DOI:10.1016/j.envpol.2016.05.021 |
Meißner K, Dittmann E, Börner T. 1996. Toxic and non-toxic strains of the cyanobacterium Microcystis aeruginosa contain sequences homologous to peptide synthetase genes. FEMS Microbiology Letters, 135(2-3): 295-303.
DOI:10.1111/j.1574-6968.1996.tb08004.x |
Mikalsen B, Boison G, Skulberg O M, Fastner J, Davies W, Gabrielsen T M, Rudi K, Jakobsen K S. 2003. Natural variation in the microcystin synthetase operon mcyABC and impact on microcystin production in Microcystis strains. Journal of Bacteriology, 185(9): 2 774-2 785.
DOI:10.1128/JB.185.9.2774-2785.2003 |
Neilan B A, Jacobs D, Therese D D, Blackall L L, Hawkins P R, Cox P T, Goodman A E. 1997. rRNA sequences and evolutionary relationships among toxic and nontoxic cyanobacteria of the genus Microcystis. International Journal of Systematic and Evolutionary Microbiology, 47(3): 693-697.
DOI:10.1099/00207713-47-3-693 |
Otsuka S, Suda S, Li R H, Matsumoto S, Watanabe M M. 2000. Morphological variability of colonies of Microcystis morphospecies in culture. The Journal of General and Applied Microbiology, 46(1): 39-50.
DOI:10.2323/jgam.46.39 |
Otsuka S, Suda S, Li R H, Watanabe M, Oyaizu H, Matsumoto S, Watanabe M M. 1998. 16S rDNA sequences and phylogenetic analyses of Microcystis strains with and without phycoerythrin. FEMS Microbiology Letters, 164(1): 119-124.
DOI:10.1111/j.1574-6968.1998.tb13076.x |
Otsuka S, Suda S, Li R H, Watanabe M, Oyaizu H, Matsumoto S, Watanabe M M. 1999a. Phylogenetic relationships between toxic and non-toxic strains of the genus Microcystis based on 16S to 23S internal transcribed spacer sequence. FEMS Microbiology Letters, 172(1): 15-21.
DOI:10.1111/j.1574-6968.1999.tb13443.x |
Otsuka S, Suda S, Li R H, Watanabe M, Oyaizu H, Matsumoto S, Watanabe M M. 1999b. Characterization of morphospecies and strains of the genus Microcystis (Cyanobacteria) for a reconsideration of species classification. Phycological Research, 47(3): 189-197.
DOI:10.1046/j.1440-1835.1999.00162.x |
Otten T G, Paerl H W. 2011. Phylogenetic inference of colony isolates comprising seasonal Microcystis blooms in Lake Taihu, China. Microbial Ecology, 62(4): 907-918.
DOI:10.1007/s00248-011-9884-x |
Palinska K A, Liesack W, Rhiel E, Krumbein W E. 1996. Phenotype variability of identical genotypes: the need for a combined approach in cyanobacterial taxonomy demonstrated on Merismopedia-like isolates. Archives of Microbiology, 166(4): 224-233.
DOI:10.1007/s002030050378 |
Pan H, Song L R, Liu Y D, Zhu Y Z, Shen Q. 2001. Characterization of toxic waterbloom-forming cyanobacteria by modified PCR. Acta Hydrobiologica Sinica, 25(2): 159-166.
(in Chinese with English abstract) DOI:10.3321/j.issn:1000-3207.2001.02.009 |
Pérez-Carrascal O M, Terrat Y, Giani A, Fortin N, Greer C W, Tromas N, Shapiro B J. 2019. Coherence of Microcystis species revealed through population genomics. The ISME Journal, 13(12): 2887-2900.
DOI:10.1038/s41396-019-0481-1 |
Rantala A, Fewer D P, Hisbergues M, Rouhiainen L, Vaitomaa J, Börner T, Sivonen K. 2004. Phylogenetic evidence for the early evolution of microcystin synthesis. Proceedings of the National Academy of Sciences of the United States of America, 101(2): 568-573.
DOI:10.1073/pnas.0304489101 |
Shan K, Shang M S, Zhou B T, Li L, Wang X X, Yang H, Song L R. 2019. Application of Bayesian network including Microcystis morphospecies for microcystin risk assessment in three cyanobacterial bloom-plagued lakes, China. Harmful Algae, 83: 14-24.
DOI:10.1016/j.hal.2019.01.005 |
Shen L Q, Ma S C, Cai F F, Yu G L, Li S C, Li R H. 2018. Polyphasic examination on Merismopedia tenuissima CHAB 7021 from Ganjiang River, China revealed the polyphyly of the genus Merismopedia (Cyanobacteria). Journal of Oceanology and Limnology, 36(4): 1 157-1 165.
DOI:10.1007/s00343-018-7341-0 |
Sidelev S, Zubishina A, Chernova E. 2020. Distribution of microcystin-producing genes in Microcystis colonies from some Russian freshwaters: is there any correlation with morphospecies and colony size?. Toxicon, 184: 136-142.
DOI:10.1016/j.toxicon.2020.06.005 |
Song L R, Sano T, Li R H, Watanabe M M, Liu Y D, Kaya K. 1998. Microcystin production of Microcystis viridis (cyanobacteria) under different culture conditions. Phycological Research, 46(S2): 19-23.
DOI:10.1046/j.1440-1835.1998.00120.x |
Tan W H, Liu Y, Wu Z X, Lin S, Yu G L, Yu B S, Li R H. 2010. CpcBA-IGS as an effective marker to characterize Microcystis wesenbergii (Komárek) Komárek in Kondrateva (cyanobacteria). Harmful Algae, 9(6): 607-612.
DOI:10.1016/j.hal.2010.04.011 |
Tanabe Y, Kaya K, Watanabe M M. 2004. Evidence for recombination in the microcystin synthetase (mcy) genes of toxic cyanobacteria Microcystis spp. Journal of Molecular Evolution, 58(6): 633-641.
DOI:10.1007/s00239-004-2583-1 |
Tsujimura S, Tsukada H, Nakahara H, Nakajima T, Nishino M. 2000. Seasonal variations of Microcystis populations in sediments of Lake Biwa, Japan. Hydrobiologia, 434(1): 183-192.
DOI:10.1023/A:1004077225916 |
Vasconcelos V M, Pereira E. 2001. Cyanobacteria diversity and toxicity in a wastewater treatment plant (Portugal). Water Research, 35(5): 1 354-1 357.
DOI:10.1016/S0043-1354(00)00512-1 |
Via-Ordorika L, Fastner J, Kurmayer R, Hisbergues M, Dittmann E, Komarek J, Erhard M, Chorus I. 2004. Distribution of microcystin-producing and non-microcystin-producing Microcystis sp.in European freshwater bodies: detection of microcystins and microcystin genes in individual colonies. Systematic and Applied Microbiology, 27(5): 592-602.
DOI:10.1078/0723202041748163 |
Wang H J, Xu C, Liu Y, Jeppesen E, Svenning J C, Wu J, Wu J G, Zhang W X, Zhou T J, Wang P, Nangombe S, Ma J, Duan H T, Fang J Y, Xie P. 2021. From unusual suspect to serial killer: cyanotoxins boosted by climate change may jeopardize megafauna. The Innovation, 2(2): 100092.
DOI:10.1016/j.xinn.2021.100092 |
Wang Z J, Song G F, Li Y G, Yu G L, Hou X Y, Gan Z X, Li R H. 2019. The diversity, origin, and evolutionary analysis of geosmin synthase gene in cyanobacteria. Science of the Total Environment, 689: 789-796.
DOI:10.1016/j.scitotenv.2019.06.468 |
WHO. 1998. Guidelines for Drinking-Water Quality, Volume 2- Health Criteria and Other Supporting Information. Addendum. 2nd edn. World Health Organization, Geneva.
|
Wiedner C, Visser P M, Fastner J, Metcalf J S, Codd G A, Mur L R. 2003. Effects of light on the microcystin content of Microcystis Strain PCC 7806. Applied and Environmental Microbiology, 69(3): 1 475-1 481.
DOI:10.1128/AEM.69.3.1475-1481.2003 |
Wu Z X, Gan N Q, Song L R. 2007. Genetic diversity: geographical distribution and toxin profiles of Microcystis strains (cyanobacteria) in China. Journal of Integrative Plant Biology, 49(3): 262-269.
DOI:10.1111/j.1744-7909.2007.00368.x |
Xiao M, Willis A, Burford M A, Li M. 2017. Review: a meta-analysis comparing cell-division and cell-adhesion in Microcystis colony formation. Harmful Algae, 67: 85-91.
DOI:10.1016/j.hal.2017.06.007 |
Xu Y, Wu Z X, Yu B S, Peng X, Yu G L, Wei Z H, Wang G X, Li R H. 2008. Non-microcystin producing Microcystis wesenbergii (Komárek) Komárek (Cyanobacteria) representing a main waterbloom-forming species in Chinese waters. Environmental Pollution, 156(1): 162-167.
DOI:10.1016/j.envpol.2007.12.027 |
 2021, Vol. 39
2021, Vol. 39