Institute of Oceanology, Chinese Academy of Sciences
Article Information
- ZHANG Shuqian, ZHANG Suping
- Tritonia iocasica sp. nov., a new tritoniid species from a seamount in the tropical Western Pacific (Heterobranchia: Nudibranchia)
- Journal of Oceanology and Limnology, 39(5): 1817-1829
- http://dx.doi.org/10.1007/s00343-021-0408-3
Article History
- Received Oct. 23, 2020
- accepted in principle Dec. 18, 2020
- accepted for publication Feb. 7, 2021
2 Center for Ocean Mega-Science, Chinese Academy of Sciences, Qingdao 266071, China
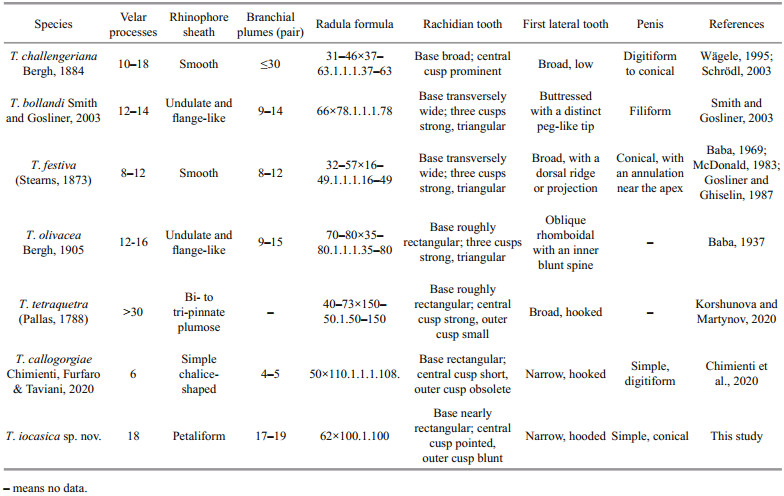
The genus Tritonia Cuvier, 1798 is a relatively diverse group of tritoniid nudibranchs that currently includes about 27 accepted species (MolluscaBase, 2021). The genus has a worldwide distribution, ranging from the Atlantic and Mediterranean (Marcus, 1983; Gosliner and Ghiselin, 1987; Silva et al., 2014; Trainito and Doneddu, 2014; Chimienti et al., 2020; Ortea and Moro, 2020), eastern Pacific (Bertsch, 2014; Valdés et al., 2018), Western Pacific (Baba, 1969; Smith and Gosliner, 2003), to Antarctica (Wägele, 1995; Schrödl, 2003; Ballesteros and Avila, 2006), from the intertidal zone to abyssal depths (2 550 m, T. nigromaculata Roginskaya, 1984). Most species are known or suspected specialist predators on octocorals, often limited to a single species or group of species of a single family (Smith and Gosliner, 2003; García-Matucheski and Muniain, 2011; Trainito and Doneddu, 2014; Chimienti et al., 2020). The genus has been relatively well studied in the Atlantic Ocean, with nearly 20 species described. In contrast, however, the fauna in the Pacific region, especially in the Western Pacific, remains poorly studied, with only four described species.
Over the recent years, increasing explorations to deep-sea benthic ecosystems have yielded several novel species to the genus Tritonia (e.g. Valdés et al., 2017, 2018; Chimienti et al., 2020; Ortea and Moro, 2020), indicating that the knowledge of deep-water fauna of the genus is still incomplete. In 2019, the Institute of Oceanology, Chinese Academy of Sciences (IOCAS) conducted a scientific cruise to a seamount at the Caroline Plate in the tropical Western Pacific. During this investigation, three individuals of a sea slug were collected. Examinations of their external morphology, radular features, and gross internal anatomy indicated that they represent an undescribed species belonging to the genus Tritonia. Examination of gut contents revealed several coral fragments and sclerites that belong to the coral genus Melithaea Milne Edwards, 1857. This is the first record of a tritoniid species feeding on corals of the family Melithaeidae Gray, 1870. Phylogenetic analyses based on two mitochondrial (COI, 16S rRNA) and a nuclear (H3) genes using Bayesian inference, maximum likelihood, and species delimitation analysis also support the placement of the new species in the genus Tritonia, and its separation from related congeners.
2 MATERIAL AND METHOD 2.1 Specimens sampling and preservationSpecimens were photographed and then collected from the upper bathyal zone (970–1 262 m) during two dives of the ROV Faxian (mother ship R/V Kexue) (IOCAS) at a seamount near the Mariana Trench (Fig. 1a). The specimens were photographed alive on board. One specimen was preserved in 99.5% ethanol (here designated as holotype MBM285105) and two others were frozen at -80 ℃ for genomic study.

|
| Fig.1 Geographical location of the type locality (a, solid red triangle) and the living specimen (paratype MBM285106) of Tritonia iocasa sp. nov. on a basalt rock at the type locality (b) |
In the course of genomic study, one of the two specimens frozen at -80 ℃ was triturated thoroughly after dissecting its jaws, radula, and digestive gland. The jaws, radula, and digestive gland, together with a photograph of the living animal, are herein designated as paratype MBM285106 as a whole. The other specimen was entirely subjected to the genomic study, with no taxonomical observation and no gene fragments (i.e. COI, 16S, and H3) sequenced.
The holotype and paratype have been deposited at the Marine Biological Museum of Chinese Academy of Sciences (MBMCAS).
2.2 Morphological examinationExternal morphology and internal anatomy were observed under a Zeiss Stemi SV 11 Apo stereomicroscope. Drawings of the digestive and reproductive systems were made with the aid of a Zeiss camera Lucida. For scanning electron microscopy (SEM) studies, jaws and radula were extracted from buccal bulb by gross dissection, cleaned using 10% NaOH, rinsed in distilled water, air-dried, coated with gold, and examined with SEM at an accelerating voltage of 5kV. Gut content and coral fragments were treated with undiluted commercial bleach in a centrifugal tube to obtain sclerites. These were washed several times with distilled water, transferred to aluminum stubs, coated with gold, and examined with Hitachi S-3400N SEM at an accelerating voltage of 5kV.
2.3 Molecular analyses 2.3.1 Extraction, amplification, and DNA sequencingHolotype and paratype (see below) were selected for molecular studies. Genomic DNA from each individual was extracted with the Column Genomic DNA Isolation Kit (Beijing TIANGEN, China) according to the manufacturer's instructions. DNA was eluted in elution buffer and stored at -20 ℃ until use. PCR reactions were carried out in a total volume of 50 μL, including 2-μL DNA template, 1.5-mmol/L MgCl2, 0.2 mmol/L of each dNTPs, 1 μL of both forward and reverse PCR primers, 10× buffer and 2.5-U Taq DNA polymerase. Thermal cycling was performed under the following conditions: 94 ℃ for 3 min (initial denaturation), followed by 35 cycles of 94 ℃ for 30 s (denaturation), 40 s at primer-specific annealing temperatures, 72 ℃ for 60 s (extension) and a final extension at 72 ℃ for 10 min. PCR and sequencing primers for COI, 16S rRNA, and H3 genes are listed in Table 1. PCR products were verified on a GelRed-stained 1.5% agarose gel and purified with the Column PCR Product Purification Kit (Shanghai Sangon, China).

|
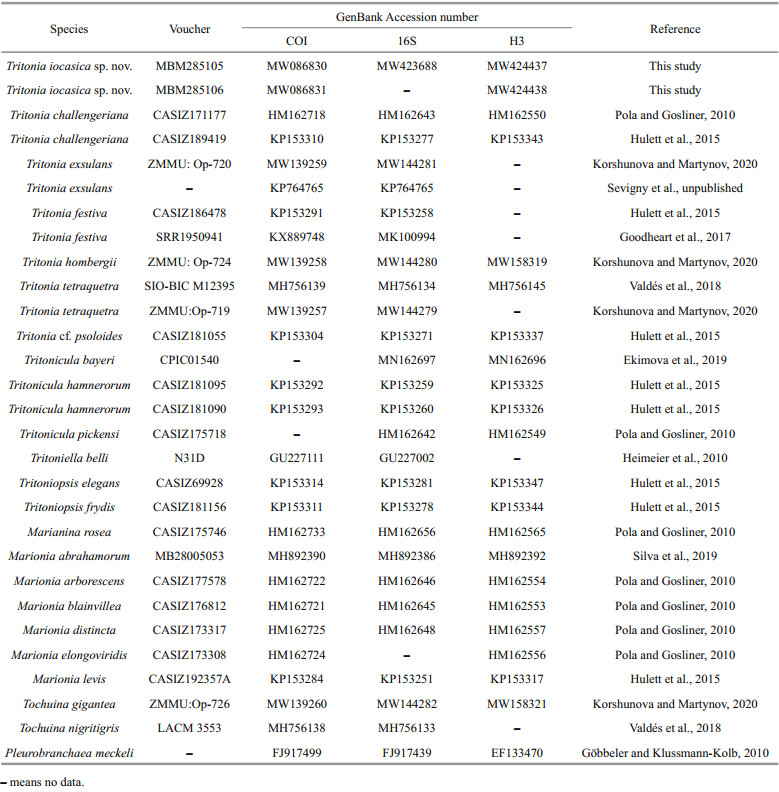
Two specimens (MBM285105 and MBM285106) were subjected to molecular analysis. In addition, other sequences of Tritonia spp. were retrieved from GenBank and used for phylogenetic analyses (Table 2). We followed Korshunova and Martynov (2020) to include other tritoniid species in the analyses for comparison, and have used Pleurobranchaea meckeli (De Blainville, 1825) to root the tree.
|
Sequence alignments were generated with the MAFFT algorithm (Katoh and Standley, 2013). COI and H3 sequences were translated into amino acids for confirmation of the alignment. Separate analyses were conducted for COI (567 bp), 16S (413 bp), H3 (288 bp), and concatenated data (1 268 bp). Ambiguously aligned fragments of 16S alignment was removed using Gblocks (Talavera and Castresana, 2007) with the following parameter settings: minimum number of sequences for a conserved/flank position (53/53), maximum number of contiguous non-conserved positions (8), minimum length of a block (10), allowed gap positions (with half). Maximum likelihood phylogenies were inferred using IQ-TREE (Nguyen et al., 2015) under Edge-unlinked partition model for 20000 ultrafast (Minh et al., 2013) bootstraps, as well as the Shimodaira-Hasegawa-like approximate likelihood-ratio test (Guindon et al., 2010). Bayesian Inference phylogenies were inferred using MrBayes 3.2.6 (Ronquist et al., 2012) under partition model (2 parallel runs, 5 000 000 generations), in which the initial 25% of sampled data were discarded as burn-in. The best-fit models of evolution (GTR+I+G for COI and 16S, GTR for H3) were determined using PartitionFinder2 (Lanfear et al., 2017), with greedy algorithm and the Akaike information criterion AIC (Akaike, 1998). The p-distances within and among each species grouping were estimated with MEGA X (Kumar et al., 2018).
To discriminate species, we included 27 COI sequences for analyses based on the Kimura 2-parameter model. Bayesian inference (BI) was conducted using the software MrBayes v. 3.2.6 (Ronquist et al., 2012) based on the GTR+I+G model. The Metropolis-coupled Monte Carlo Markov chains were run for 5 000 000 generations, with sampling every 100 generations; split frequencies were less than 0.01 before the analyses were terminated. A 25% burn-in was applied before constructing the majority-rule consensus tree. Results were visualized using FigTree V. 1.4.3.
2.4 Species delimitation analysisThe Automatic Barcode Gap Discovery (ABGD) method (Puillandre et al., 2012) was used to assess the number of Tritonia species studied herein. The alignment from the fast-evolving COI gene was uploaded to the online server of ABGD. The analysis was performed with the model of Jukes-Cantor (JC69) with the following settings (Pmin=0.001, Pmax=0.1, steps=10, X=1.0, Nb bins=20).
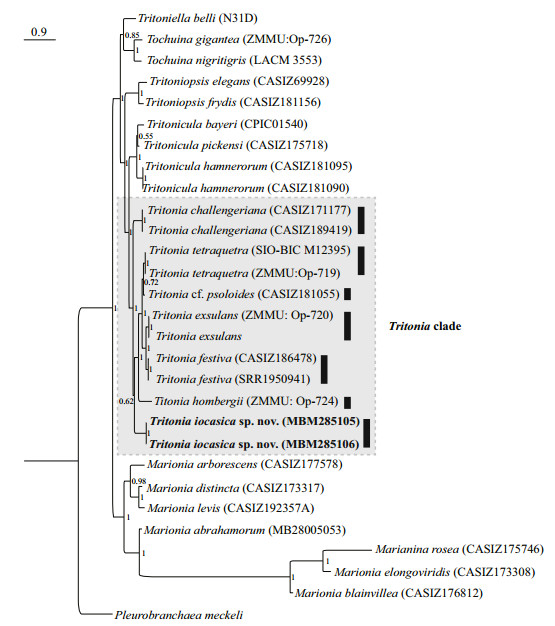
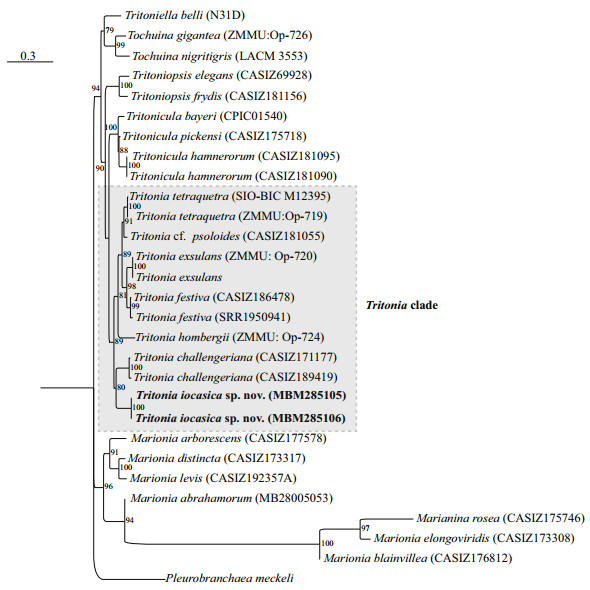
3 RESULT 3.1 Phylogenetic analysesPartial sequences (COI and H3) were obtained from the holotype (MBM285105) and paratype (MBM285106), while that of 16S was from the holotype only (see Table 2 for accession numbers). The two consensus trees inferred using Bayesian inference (BI) and maximum likelihood (ML) criteria were generally congruent (Figs. 2 & 3). The genus Tritonia, including the new species, was recovered as monophyletic with strong support (posterior probability, PP=1; bootstrap value, BS=89). The analyses based on concatenated dataset (COI, 16S, and H3) showed that T. iocasica sp. nov. form a separate clade within the genus.
|
| Fig.2 Phylogenetic tree inferred by Bayesian analysis (BI) based on concatenated dataset of COI, 16S, and H3 genes Numbers adjacent to nodes refer to BI posterior probability (PP>0.5). PP≥90 was considered as enough support. Vertical bars indicate th e results of ABGD species delimitation. Name of the new species are marked in bold. |
|
| Fig.3 Phylogenetic tree inferred by maximum likelihood (ML) based on concatenated dataset of COI, 16S, and H3 genes Numbers adjacent to nodes refer to ML bootstrap scores (BS>50). BS≥70 was considered as enough support. Name of the new species are marked in bold. |
The BI analysis based on the concatenated COI, 16S and H3 markers does not solve (PP=0.62) the relationships among T. iocasica sp. nov., T. challengeriana, and the remaining Tritonia spp. included in this study, although the monophyly of the genus is recovered. The phylogenetic tree inferred using BI criteria based on single gene COI (Supplementary Fig.S1) only support a joint relationship of T. iocasica sp. nov and T. challengeriana, but does not recover the monophyly of Tritonia.
On the other hand, the ML analysis based on the concatenated COI, 16S, and H3 markers support a joint relationship among T. iocasica sp. nov. with T. challengeriana. These two species clustered together form a clade sister to other Tritionia spp. (BS=89).
These results support the systematic placement of the new species in the genus Tritonia and its separation from other congeners.
3.2 Species delimitation analysesThe ABGD analysis of the COI sequences resulted in the delimitation of seven species (Fig. 2), with values of prior intraspecific divergence (P) being ≥0.001 0. These groups correspond to clades recovered by BI and ML analyses. Within the genus, the two specimens of the new species are closest related to each other, with 0.3% pairwise distance; with the sequences studied herein, the pairwise distances among T. iocasica sp. nov. and other species of Tritonia are 17.8%–20.5% (Table 3).
|
These results provide additional support for the separation of T. iocasica sp. nov. from its congeners.
3.3 SystematicsSuperfamily Tritonioidea Lamarck, 1809
Family Tritoniidae Lamarck, 1809
Genus Tritonia Cuvier, 1797
Type species: Tritonia hombergii Cuvier, 1803, subsequent designation by Lemehe (1961).
Tritonia iocasica sp. nov. (Figs. 1b, 4–6)
|
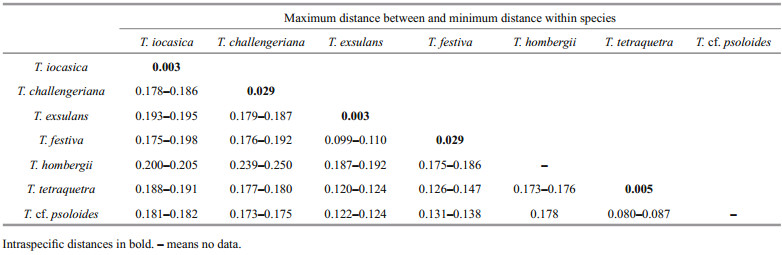
| Fig.4 Tritonia iocasica sp. nov. a–e & g. from the paratype (MBM285106); f. from the holotype (MBM285105) (a. fresh specimen on board, dorsal view; b. dorsal view of the velum; c. right view of anterior end; d. left view of anterior end, with anterior part of digestive tract evaginated; e. ventral view of partial visceral mass; f. rhinophores; g. jaws). Abbreviation: an: anus; bb: buccal bulb; dg: digestive gland; o: ovotestis; ro: reproductive opening; st: stomach. |
|
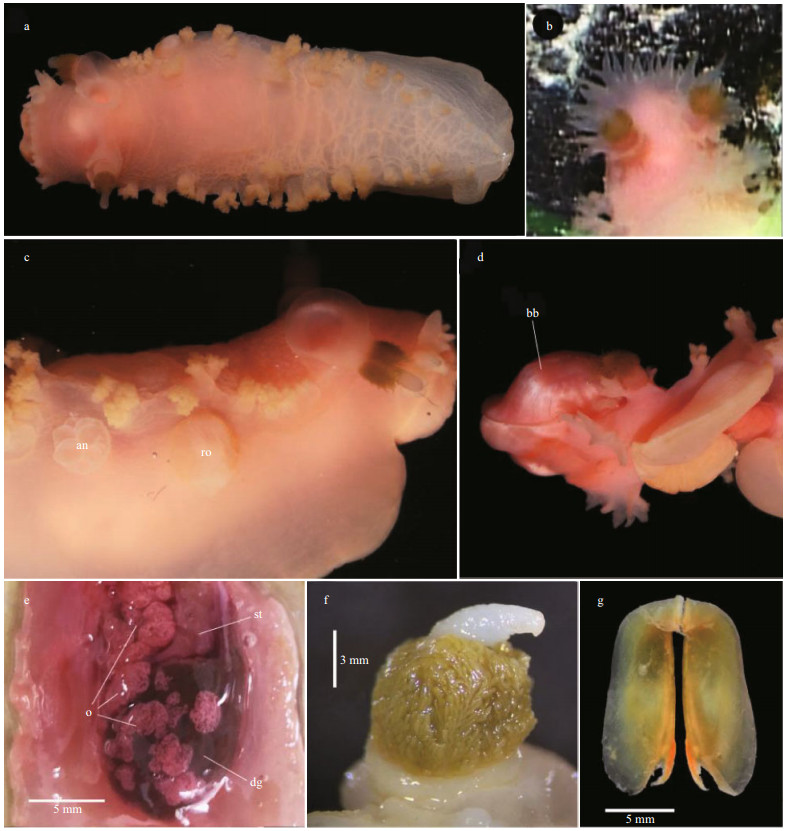
| Fig.5 Tritonia iocasica sp. nov., holotype, MBM285105 a. digestive system; b. reproductive system. Abbreviations: an: anus; am: ampulla; bb: buccal bulb; bc: bursa copulatrix; cns: central nervous system; dg: digestive gland; fgm: female gland mass; hd: hermaphrodite duct; i: intestine; p: penis; pr: prostate; sg: salivary gland; st: stomach; vg: vagina. |
|
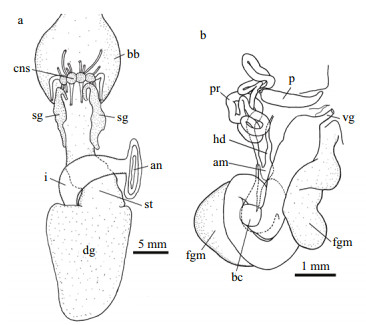
| Fig.6 Micrographs of jaw and radula of Tritonia iocasica sp. nov., specimen MBM285106 a. left jaw; b, c. jaw masticatory border; d. general view of the left part of radula; e. rachidian and inner lateral teeth; f. rachidian and the first lateral teeth; g. middle lateral teeth; h. outermost lateral teeth. |
http://zoobank.org/urn:lsid:zoobank.org:act:C29C82DB-08A5-490C-8512-3976CCD28132
Material examined: Holotype MBM285105, live length 120 mm, dissected and sequenced, Station FX-Dive 212, (10°03'N 140°11'E) at depth of 1 262-m, 30 May 2019.
Paratype MBM285106, live length 120 mm, partly used for genomic analysis, with only jaws, radula, and digestive gland remaining for taxonomical study, Station FX-Dive 209, (11°36'N, 140°12'E) at depth of 970-m, 27 May 2019.
An additional specimen, live length 110 mm, entirely used for the genomic study, with no taxonomical observation and no gene fragments sequenced. Collected together with the paratype.
Type locality: A seamount at the Caroline Plate near the Mariana Trench, 10°03'N, 140°11'E.
Etymology: The new species is named after IOCAS (Institute of Oceanology, Chinese Academy of Sciences), in celebration of its 70th anniversary.
Description:
EXTERNAL ANATOMY (Figs. 1b, 4a–c, paratype MBM285106): Body elongate, slender. Dorsum covered with weak reticulate pattern of low ridges, giving dorsal surface a fish-scale appearance. Velum rounded, with about 18 elongate digitiform velar tentacles (Fig. 4b). Notal margin with 17–19 pairs of secondary gills. Rhinophores long, located on anterior end of notum, bearing a cluster of pinnate processes that surrounds cylindrical apical papilla. Rhinophoral sheath wide, divided into several (~10) petaliform lobes. Foot smooth, anterior end rounded, gradually tapering posteriorly. Genital, anal opening on the right side of body (Fig. 4c). Anus situated below the 5th and 6th secondary gills, genital opening below 3rd secondary gills. Background color of living animal uniformly pinkish (same color as prey) except for pinnate processes on rhinophores which is brownish. Lighter pink on posterior 2/3 of the notum, secondary gills, and velum.
INTERNAL ANATOMY: Digestive system (Fig. 5a, holotype MBM285105) with thick, muscular buccal bulb (Figs. 4d & 5a). Esophagus wide, anterior part connecting into buccal bulb. Posteriorly, it expands into a wider tube connecting to the stomach. The stomach oval in shape and its posterior part embedded ventrally into the digestive gland. Two long salivary glands connecting with the buccal bulb at each side of the esophageal junction. Intestine emerging dorsally from left side of digestive gland, forming simple loop towards right side of body, where it opens into the anus. Stomach simple, no stomach plates present. Digestive gland reddish brown, large (Fig. 4e). Jaws (Figs. 4g, 6a–c, paratype MBM285106) thin, elongate, amber-colored, inner margin (masticatory margin) terminating distally in small pointed process. The masticatory margin apparently worn at upper end; 1/3 lower part with about 18 or more rows of rodlets. Maximum length of rodlets about 200 μm in length, and maximum width about 40 μm. Most rodlets with irregularly pentagonal or hexagonal cross section. Radula (Fig. 6d–h, paratype MBM285106) with a formula of 62×100.1.100. Rachidian teeth tricuspid, with nearly rectangular bases. Central cusp sharply pointed and longer; outer cusps blunt and shorter, fused to margin of base. Distinct, deep incision between central cusp and right outer cusp (Fig. 6f). First lateral teeth with thicker base, sturdy, hoodlike cusps (Fig. 6g). Remaining lateral teeth sickle-shaped, with sharp cusps, becoming thin towards outer margins of radula (Fig. 6h).
Reproductive system (Fig. 5b, holotype MBM285105) androdiaulic. Ovotestis covering most of surface of digestive gland, uniformly pinkish (Fig. 4e). Ampulla short, simply convoluted, opening on female gland mass where prostate connects. Prostate narrow, elongate, forming several loops and ending into narrow deferent duct, which connects with conical penial sac. Penis simple, elongate. Vagina elongate, broad, opening into the spherical bursa copulatrix.
Diet: Intestinal tract contained coral fragments and calcareous sclerites (Fig. 7) of an unidentified coral species, Melithaea sp. (Melithaeidae).

|
| Fig.7 Coral fragments belonging to Melithaea sp. dissected from intestinal tract of specimen MBM285106 (a) and different forms of sclerites of coral (b) |
The systematics of Tritoniidae has long been considered problematic despite the revisions by Odhner(1936, 1963) and Marcus (1983). Relationships among the genera are still unclear and the placement of some species remain tentative (Willan, 1988; Schrödl, 2003). The fact that previous authors utilized different characters to distinguish genera may account for the problem. Nevertheless, recent phylogenetic studies also showed that most tritoniid genera currently recognized are not monophyletic (Bertsch et al., 2009; Almón et al., 2018). A revision of the whole family based on more detailed anatomical information and molecular evidence is therefore needed to define generic boundaries (Korshunova and Martynov, 2020). Gosliner and Ghiselin (1987) treated all species of tritoniids with well developed secondary gills, with a tricuspid rachidian tooth, and without stomach plates as members of Tritonia. In the present study, we follow this classification, to assign the new species to the genus Tritonia. This assignation was supported by molecular evidence. In the phylogenetic tree reconstructed herein (Figs. 2 & 3), T. hombergii, the type species of the genus, together with other Tritonia spp., was recovered as a monophyletic clade (PP=1, BS=89). The trees based on concatenated data and single COI gene all showed that T. iocasica sp. nov. form a separate clade sister to other Tritonia spp. The species delimitation and p-distance analyses strongly support the separation of T. iocasica sp. nov. from its congeners. Within the genus, T. iocasica sp. nov. is unique by its external morphology and distinct color pattern. At present, no other known species, to our knowledge, can be confused with the new species. In the Western Pacific region, there are only four other Tritonia species that have been described (Table 4), including T. bollandi Smith and Gosliner, 2003 from Okinawa, Japan and Indonesia (58-88-m depth), T. festiva (Stearns, 1873), an intertidal species native to Pacific coast of North America but also occurs in Japan Sea (Baba, 1969), T. olivacea Bergh, 1905 from Indonesia (32-m depth) and T. tetraquetra (Pallas, 1788) from Northwestern Pacific (1-700-m depth). T. iocasica sp. nov. thus represents the first of Tritonia known from deep sea of the tropical Western Pacific, and, to our knowledge, the first for the family Tritoniidae. Unlike in the Atlantic and the eastern Pacific, the taxonomic knowledge of the genus in the Western Pacific is very poor, although several additional undescribed species were reported and illustrated by Gosliner et al.(2008, 2018). This probably results from the fact that deep-sea fauna of the Western Pacific was relatively poorly investigated. Another explanation is that many recorded Tritonia species were unidentified or misidentified, as previous taxonomic studies were based primarily on few external characters that always result in taxonomic confusions and in turn, an inaccurate estimate of biodiversity. In central Pacific, an individual likely belonging to the genus was photographed by ROV Deep Discoverer (NOAA) from Musicians Seamounts (see https://www.ncei.noaa.gov/waf/okeanos-animal-guide/Gastropoda030.html). This individual is highly similar to T. iocasica sp. nov. in coloration pattern, body shape, numbers of secondary gills and velar tentacles, and thus may be conspecific with the new species described herein. If this is the case, it would extends the distribution of T. iocasica sp. nov. to the central Pacific.
Internally, T. iocasica sp. nov. has a jaw armed with multiple rows of rodlets (Fig. 5b & c). This type of denticulation has been only recorded for a single species in the genus (Chimienti et al., 2020). In the whole family, only two other species, Marionia bathycarolinensis Smith and Gosliner, 2005 and Marionia tedi Ev. Marcus, 1983, were known to have a jaw armed with rows of rodlets (Smith and Gosliner, 2005; Valdés, 2006). Compared to these three species, the rodlets in the new species is much longer. The type of this denticulation may facilitate the distribution of stress that might damage the thin, delicate inner edge of the jaw during cutting the coral stem.
Examination of the digestive tract of T. iocasica sp. nov. revealed that the gut contains several coral fragments and calcareous sclerites that belong to Melithaea sp. The three specimens studied herein were collected from a single seamount near the Mariana Trench during two ROV dives. Two of the specimens (Fig. 1b) were discovered crawling on a platform of basalt rock. The entire area is partially colonized by primnoid octocorals, flabelliid hexacorals, the hydroid Stegolaria sp., and pheronematid sponges. Two or three groups of Melithaea sp. (Melithaeidae) were observed (not collected) from distance of about 1.5–2 m. The diet of the Tritonia species was reviewed by García-Matucheski and Muniain (2011); the members exclusively feed on octocorals, especially on the gorgonians, often limited to a single species or group of species within a single family. The present study represents the first record of predation by a tritoniid species on octocoral of family Melithaeidae, which expands our knowledge of the predation range of Tritonia.
The majority of Tritonia species live in intertidal or shallow subtidal waters, and only five have been reported down to 500 m (Bergh, 1899; Bouchet, 1977; Roginskaya, 1984; Valdés et al., 2017, 2018). Until now, the evolutionary history of species in Tritonia is still little known. Actually, it is difficult to make speculation as to the time of the Tritonia species originated from shallow water or deep sea, due to lack of a fossil record for these shell-less mollusks.
5 DATA AVAILABILITY STATEMENTThe authors declare that all the data supporting the findings of this study are available within the article.
6 ACKNOWLEDGMENTThe data and samples were collected onboard R/V Kexue. We would like to express our sincere thanks to the crews for their assistance during the survey. We are very grateful to Prof. Kuidong XU and Dr. Xuwen WU for their efforts in collecting the specimens. Dr. Yang LI identified the coral and helped in conducting the SEM study of the coral sclerites. Four anonymous reviewers provided valuable comments and suggestions that greatly improved the manuscript.
Electronic supplementary materialSupplementary material (Supplementary Fig.S1) is available in the online version of this article at https://doi.org/10.1007/s00343-021-0408-3.
Akaike H. 1998. Information theory and an extension of the maximum likelihood principle. In: Parzen E, Tanabe K, Kitagawa G eds. Selected Papers of Hirotugu Akaike. Springer, New York. p. 199–213, https://doi.org/10.1007/978-1-4612-1694-0_15.
|
Almón B, Pérez J, Caballer M. 2018. Expect the unexpected: a new large species of Marionia (Heterobranchia: nudibranchia: tritoniidae) from western Europe. Invertebrate Systematics, 32(4): 890-906.
DOI:10.1071/IS17073 |
Baba K. 1937. A new species of the nudibranchiate genus Marionia from Sagami Bay, Japan. The Venus, 7(3): 116-120.
|
Baba K. 1969. Notes on the collection of Tritonia festiva (Stearns, 1873) from the Seas of Japan (Gastropoda: Nudibranchia). The Veliger, 12: 132-134.
|
Ballesteros M, Avila C. 2006. A new tritoniid species (Mollusca: opisthobranchia) from Bouvet Island. Polar Biology, 29(2): 128-136.
DOI:10.1007/s00300,005-0059-4 |
Bergh R. 1899. Nudibranchiate Gasteropoda. Dan Ingolf Exped, 2: 1-49.
|
Bertsch H, Valdés Á, Gosliner T M. 2009. A new species of tritoniid nudibranch, the first found feeding on a zoanthid anthozoan, with a preliminary phylogeny of the Tritoniidae. Proceedings of the California Academy of Sciences, 60(12): 431-446.
|
Bertsch H. 2014. Biodiversity in La Reserva de la Biósfera Bahía de los Ángeles y Canales de Ballenas y Salsipuedes: naming of a new genus, range extensions and new records, and species list of Heterobranchia (Mollusca: gastropoda), with comments on biodiversity conservation within Marine Reserves. The Festivus, 46(5): 158-177.
|
Bouchet P. 1977. Opisthobranches de profondeur de l'Océan Atlantique: II—Notaspidea et Nudibranchiata. Journal of Molluscan Studies, 43(1): 28-66.
|
Chimienti G, Angeletti L, Furfaro G, Canese S, Taviani M. 2020. Habitat, morphology and trophism of Tritonia callogorgiae sp. nov., a large nudibranch inhabiting Callogorgia verticillata forests in the Mediterranean Sea. Deep Sea Research Part I: Oceanographic Research Papers, 165: 103364.
DOI:10.1016/j.dsr.2020.103364 |
Colgan D J, McLauchlan A, Wilson G D F, Livingston S P, Edgecombe G D, Macaranas J, Cassis G, Gray M R. 1998. Histone H3 and U2 snRNA DNA sequences and arthropod molecular evolution. Australian Journal of Zoology, 46(5): 419-437.
DOI:10.1071/ZO98048 |
Ekimova I, Valdés Á, Chichvarkhin A, Antokhina T, Lindsay T, Schepetov D. 2019. Diet-driven ecological radiation and allopatric speciation result in high species diversity in a temperate-cold water marine genus Dendronotus (Gastropoda: nudibranchia). Molecular Phylogenetics and Evolution, 141: 106609.
DOI:10.1016/j.ympev.2019.106609 |
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology, 3(5): 294-299.
|
García-Matucheski S, Muniain C. 2011. Predation by the nudibranch Tritonia odhneri (Opisthobranchia: tritoniidae) on octocorals from the South Atlantic Ocean. Marine Biodiversity, 41(2): 287-297.
DOI:10.1007/s12526-010-0063-y |
Göbbeler K, Klussmann-Kolb A. 2010. Out of Antarctica?- new insights into the phylogeny and biogeography of the Pleurobranchomorpha (Mollusca, Gastropoda). Molecular Phylogenetics and Evolution, 55(3): 996-1.
DOI:10.1016/j.ympev.2009.11.027 |
Goodheart J A, Bazinet A L, Valdés Á, Collins A G, Cummings M P. 2017. Prey preference follows phylogeny: evolutionary dietary patterns within the marine gastropod group Cladobranchia (Gastropoda: heterobranchia: nudibranchia). BMC Evolutionary Biology, 17(1): 221.
DOI:10.1186/s12862-017-1066-0 |
Gosliner T M, Behrens D W, Valdés Á. 2008. Indo-Pacific Nudibranchs and Sea Slugs: a Field Guide to the World's Most Diverse Fauna. California Academy of Sciences/Sea Challengers Natural History Books, San Francisco. 426p.
|
Gosliner T M, Ghiselin M T. 1987. A new species of Tritonia (Opisthobranchia: gastropoda) from the Caribbean Sea. Bulletin of Marine Science, 40(3): 428-436.
|
Gosliner T M, Valdés Á, Behrens D W. 2018. Nudibranch & Sea Slug Identification: Indo-Pacific. 2nd ed. New World Publications, Jacksonville. 451p.
|
Guindon S, Dufayard J F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology, 59(3): 307-321.
DOI:10.1093/sysbio/syq010 |
Heimeier D, Lavery S, Sewell M A. 2010. Using DNA barcoding and phylogenetics to identify Antarctic invertebrate larvae: lessons from a large scale study. Marine Genomics, 3(3-4): 165-177.
DOI:10.1016/j.margen.2010.09.004 |
Hulett R E, Mahguib J, Gosliner T M, Valdés Á. 2015. Molecular evaluation of the phylogenetic position of the enigmatic species Trivettea papalotla (Bertsch) (Mollusca: nudibranchia). Invertebrate Systematics, 29(3): 215-222.
DOI:10.1071/IS15002 |
Katoh K, Standley D M. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution, 30(4): 772-780.
DOI:10.1093/molbev/mst010 |
Korshunova T, Martynov A. 2020. Consolidated data on the phylogeny and evolution of the family Tritoniidae (Gastropoda: nudibranchia) contribute to genera reassessment and clarify the taxonomic status of the neuroscience models Tritonia and Tochuina. PLoS One, 15(11): e0242103.
DOI:10.1371/journal.pone.0242103 |
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35(6): 1547-1549.
DOI:10.1093/molbev/msy096 |
Lanfear R, Frandsen P B, Wright A M, Senfeld T, Calcott B. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution, 34(3): 772-773.
DOI:10.1093/molbev/msw260 |
Lemehe H. 1961. Tritonia Cuvier, [1797] (Gastropoda); proposed designation of a type-species under the plenary powers. The Bulletin of Zoological Nomenclature, 18: 285-286.
|
Marcus E D B R. 1983. The western Atlantic Tritoniidae. Boletim de Zoologia, 6(6): 177-214.
DOI:10.11606/issn.2526-3358.bolzoo.1983.121960 |
McDonald G R. 1983. A review of the nudibranchs of the California coast. Malacologia, 24(1-2): 114-276.
|
Minh B Q, Nguyen M A T, Von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Molecular Biology and Evolution, 30(5): 1188-1195.
DOI:10.1093/molbev/mst024 |
MolluscaBase. 2021. MolluscaBase. Tritonia Cuvier, 1798. Accessed through: World Register of Marine Species. http://www.marinespecies.org/aphia.php?p=taxdetails&id=138580, accessed on 2021-06-08.
|
Nguyen L T, Schmidt H A, Von Haeseler A, Minh B Q. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution, 32(1): 268-274.
DOI:10.1093/molbev/msu300 |
Odhner N H. 1936. Nudibranchia dendronotacea: a revision of the system. In: Mélanges P P ed. Mémoires du Musée Royal d'Histoire Naturelle de Belgique. Musée Royal d'Histoire Naturelle de Belgique, Bruxelles. p. 1057–1128.
|
Odhner N H. 1963. On the taxonomy of the family Tritoniidae (Mollusca: opisthobranchia). The Veliger, 6: 48-52.
|
Ortea J, Moro L. 2020. El FS Poseidon rinde homenaje al Hirondelle: descripción de una nueva especie de Tritonia Cuvier, 1798 (Mollusca: nudibranchia) de las aguas profundas de las islas de Cabo Verde para celebrar las singladuras de un yate carismático. Avicennia, 27: 29-34.
|
Palumbi S R. 1996. Nucleic acids II: the polymerase chain reaction. In: Hillis D M, Moritz C, Mable B K eds. Molecular Systematics. Sinauer Associates, Sunderland, Massachusetts. p. 205–247.
|
Pola M, Gosliner T M. 2010. The first molecular phylogeny of cladobranchian opisthobranchs (Mollusca, Gastropoda, Nudibranchia). Molecular Phylogenetics and Evolution, 56(3): 931-941.
DOI:10.1016/j.ympev.2010.05.003 |
Puillandre N, Lambert A, Brouillet S, Achaz G. 2012. ABGD, automatic barcode gap discovery for primary species delimitation. Molecular Ecology, 21(8): 1864-1877.
DOI:10.1111/j.1365-294x.2011.05239.x |
Roginskaya I S. 1984. A new deep-water species of Tritoniidae (Opisthobranchia, Nudibranchia, Dendronotacea) from the slope of Kurile Islands. Trudy Instituta Okeanologii Akademii Nauk SSSR, 119: 99-105.
|
Ronquist F, Teslenko M, Van Der Mark P, Ayres D L, Darling A, Höhna S, Larget B, Liu L, Suchard M A, Huelsenbeck J P. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61(3): 539-542.
DOI:10.1093/sysbio/sys029 |
Schrödl M. 2003. Sea slugs of South America: Systematics, Biogeography and Biology of Chilean and Magellanic Nudipleura (Mollusca: Opisthobranchia). ConchBooks, Hackenheim. 165p.
|
Silva F D V, De Azevedo V M, Matthews-Cascon H. 2014. A new species of Tritonia (Opisthobranchia: nudibranchia: tritoniidae) from the tropical South Atlantic Ocean. Journal of the Marine Biological Association of the United Kingdom, 94(3): 579-585.
DOI:10.1017/S0025315413001586 |
Silva F D V, Herrero-Barrencua A, Pola M, Cervera J L. 2019. Description of a new species of Marionia (Gastropoda: heterobranchia: tritoniidae) from the Gulf of Guinea (eastern Atlantic Ocean). Bulletin of Marine Science, 95(3): 431-447.
DOI:10.5343/bms.2018.0096 |
Smith V G, Gosliner T M. 2003. A new species of Tritonia from Okinawa (Mollusca: nudibranchia), and its association with a gorgonian octocoral. Proceedings of the California Academy of Sciences, 54(16): 255-278.
|
Smith V G, Gosliner T M. 2005. A new species of Marionia (Gastropoda: nudibranchia) from the Caroline Islands. Proceedings of the California Academy of Sciences, 56(6): 66-75.
|
Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Systematic Biology, 56(4): 564-577.
DOI:10.1080/10635150701472164 |
Trainito E, Doneddu M. 2014. Nudibranchi del Mediterraneo. Il Castello, Milano. 192p.
|
Valdés Á, Lundsten L, Wilson N G. 2018. Five new deep-sea species of nudibranchs (Gastropoda: heterobranchia: cladobranchia) from the Northeast Pacific. Zootaxa, 4526(4): 401433.
DOI:10.11646/zootaxa.4526.4.1 |
Valdés Á, Murillo F J, McCarthy J B, Yedinak N. 2017. New deep-water records and species of North Atlantic nudibranchs (Mollusca, Gastropoda: heterobranchia) with the description of a new species. Journal of the Marine Biological Association of the United Kingdom, 97(2): 303-319.
DOI:10.1017/S0025315416000394 |
Valdés A. 2006. Marionia tedi Ev. Marcus, 1983 (Nudibranchia, Tritoniidae) in the Gulf of Mexico: first record of an opisthobranch mollusk from hydrocarbon cold seeps. Gulf and Caribbean Research, 18(1): 41-46.
DOI:10.18785/gcr.1801.05 |
Wägele H. 1995. The morphology and taxonomy of the Antarctic species of Tritonia Cuvier, 1797 (Nudibranchia: dendronotoidea). Zoological Journal of the Linnean Society, 113(1): 21-46.
DOI:10.1016/S0024,4082(05)80003-2 |
Willan R C. 1988. The taxonomy of two host-specific, cryptic dendronotoid nudibranch species (Mollusca: gastropoda) from Australia including a new species description. Zoological Journal of the Linnean Society, 94(1): 39-63.
DOI:10.1111/j.1096-3642.1988.tb00881.x |
 2021, Vol. 39
2021, Vol. 39