Institute of Oceanology, Chinese Academy of Sciences
Article Information
- YANG Ya'nan, CUI Zhaoxia, FENG Tianyi, BAO Chenchang, XU Yuanfeng
- Transcriptome analysis elucidates key changes of pleon in the process of carcinization
- Journal of Oceanology and Limnology, 39(4): 1471-1484
- http://dx.doi.org/10.1007/s00343-020-0176-5
Article History
- Received Apr. 27, 2020
- accepted in principle Jun. 8, 2020
- accepted for publication Aug. 24, 2021
Metamorphosis is an important developmental transition in a large number of animal taxa. Metamorphosis refers to the process in which an immature individual undergoes drastic anatomical and physiological changes to develop into an adult in the life cycle (Medina, 2009). In the brachyuran decapod, crabs go through a carcinization morphogenesis process, involving remarkable morphological and anatomical transformations. In freshwater crabs, these occur embyonically, since all true freshwater crabs lack free-swimming larval stages and exhibit direct development (Davie et al., 2015). Due to carcinization, crabs have the shortened and compressed pleon, folded beneath the cephalothorax, and inserted between the pereiopods or in a special cavity (McLaughlin and Lemaitre, 1997; Guinot and Bouchard, 1998; Scholtz, 2014), the degeneration of pleopods (Lee et al., 1994), and the fusion of ventral nerve cords (Davie et al., 2015). In some crabs, the secondary sexual characteristics appear in this process, including the formation of gonopores and gonopods (Payen, 1975; Lee et al., 1994; Davie et al., 2015).
The Chinese mitten crab, Eriocheir sinensis is one of the most commercially important aquacultural species in China (FAO, 2020). Thanks to the development of hatchery and larviculture techniques, larvae are available from captive cultures, and aquaculture is not reliant on wild sources; as a result, the Chinese mitten crab aquaculture spreads nearly all over China (Sui et al., 2011). The larval development of E. sinensis includes five zoeal stages and one megalopa stage before the megalopa molts to the first juvenile crab stage (Anger, 1991). When the megalopa molting to the first juvenile crab stage, the crabs undergo carcinization morphogenesis, such as the reduction and folding of the pleon beneath the cephalothorax (Guinot and Bouchard, 1998) and the fusion of ventral nerve cord and thoracic ganglion (Song et al., 2017). To date, carcinization of E. sinensis has been studied at a transcriptomic level (Song et al., 2015), and some genes such as proliferating cell nuclear antigen (PCNA), cuticle and ferritin had been identified in this process (Li et al., 2011). It remains unclear whether the described genes are related to the key physiological and morphological processes in carcinization morphogenesis.
To study the molecular mechanism of pleon metamorphogenesis during carcinization, we performed a comparative transcriptomic analysis for the cephalothoraxes and pleons of the megalopa and the first juvenile crab stage. Additionally, we performed an enrichment analysis on the key physiological and morphological changes, including energy metabolism, morphological changes of pleon, and ventral nerve cord fusion. Specifically, our analysis focused on how the Hox genes, abdominal-A (abd-A) and abdominal-B (abd-B), regulate the carcinization. Researching the regulatory mechanisms of carcinization process may help us better understand the developmental biology of E. sinensis.
2 MATERIAL AND METHOD 2.1 Sample preparation and morphological observation of pleonsMegalopaes (M) and the first juvenile crab stage (J) of E. sinensis were collected from a farm in Nantong, Jiangsu province, China. For molecular studies, the cephalothoraxes and pleons were frozen in liquid nitrogen immediately. Three biological replicates were performed in cephalothoraxes of megalopaes (MC) and the first juvenile crab stage (JC), and in pleons of megalopaes (MP) and the first juvenile crab stage (JP) respectively, receiving a total of 12 samples.
For morphological observations, the external characteristics (pleopods and pleon shape) of exuviaes from megalopae (n=20) and whole body from the first juvenile crab stage (n=20), were photographed using Olympus SZ51 microscopes.
2.2 cDNA library construction and sequencingTotal RNA was isolated with the Trizol Reagent (Invitrogen, USA) according to the manufacturer's instructions. RNA quality and concentration were determined using a Qubit® RNA Assay Kit (Life Technologies, CA, USA) and a NanoDrop 6000 (Agilent Technologies, Palo Alto, CA, USA). mRNA poly (A) was separated using oligo (dT) beads, and then digested into short fragments with a TruSeq RNA Sample Prep Kit (Illumina, San Diego, CA, USA). Double strands cDNA were synthesized with random hexamer primers. The cDNA fragment and sequencing adaptor were ligated with T4 DNA ligase according to Illumina manufacturer's protocol. The ligated products were amplified to create a cDNA library. Each cDNA library was sequenced by the Illumina HiSeqTM 2000 platform.
2.3 Transcriptome assembly and annotationClean reads were guaranteed by removing adapter, filtering low-quality sequences (< Q20) and sequences shorter than 50 bp. The clean reads were assembled using Trinity software (http://trinityrnaseq.sourceforge.net/) as described for de novo transcriptome assembly without a reference genome (Garber et al., 2011).
Gene functional annotations were performed basing on public databases. Each transcript was compared using Blastx algorithm (E≤1e-5) with the National Center for Biotechnology Information (NCBI) nonredundant protein sequence (Nr) database (http://www.ncbi.nlm.nih.gov/) and clustered into a unique sequence (unigene). Next, Hmmscan (HMMER 3.0) was used to search for Protein family (Pfam) domain signatures in unigenes. Moreover, animal transcription factors (TFs) database AnimalTFDB version 2.0 (http://bioinfo.life.hust.edu.cn/AnimalTFDB/) was used to identify TFs. Gene Ontology (GO) (http://www.geneontology.org/) annotations and functional classification of all unigenes were obtained by using Blast2GO. The biochemical pathways were predicted by Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg/). Furthermore, NCBI nucleotide sequences database (Nt), SwissProt database, and eukaryotic Orhtolog Group database (KOG) were also used to annotate unigenes.
2.4 Gene expression analysisTo detect the expression levels of each unigene, all clean reads in each sample were first aligned with Unigene database using Bowtie (Langmead, 2010) before performing the estimation of expression level by combined RNA-Seq by expectation maximization (RSEM) (Li and Dewey, 2011). The fragments per kilobase of transcript sequence per million mapped reads (FPKM) (Trapnell et al., 2010)value was used to estimate the unigene expression abundance.
2.5 Identification and validation of differentially expressed genesDifferentially expressed genes (DEGs) between two transcriptomes were identified by the DESeq program (Love et al., 2014). DEGs were screened out according to the difference of expression amount fold change (FC≥2) and significant P-value (Padj < 0.05). GO enrichment analysis of DEGs was conducted by using the topGO software. KOG statistical classification and KEGG annotation of DEGs were also performed. The significance of enriched pathway was analyzed using the method of Fisher's exact test (Tavazoie et al., 1999). Functional domain analysis was performed with ExPASy PROSITE (http://www.expasy.ch/tools/scanprosite/).
Total RNA were isolated from MC, MP, JC, and JP as described above. Approximately 2-μg RNA were used to the reverse transcription of cDNA. The fluorescent quantitative real-time PCR (qRT-PCR) was used to validate the expressed transcripts of DEGs. DEGs in Hedgehog and Hippo pathways included abd-A, abd-B, p53, and Dachs. Meanwhile, the randomly selected genes included hypothetical protein T11_4388 (HP), prophenoloxidase-activating factor (PPAF), fatty acid binding protein (FABP), crustin, hemocyanin subunit 6 (HS) and cuticle proprotein proCP2.2 (CP). Gene specific primers were designed according to the Illumina sequencing data and the housekeeping gene β-actin was used as an internal control (Supplementary Table S1). The qRT-PCR was carried out with 2×SYBR Select Master Mix (Applied Biosystems, Austin, TX, USA) as the manufacturer's instructions in a 7500 Real-time PCR System (Applied Biosystems, Carlsbad, CA USA). All experiments were performed in triplicate. Gene expression level relative to control was determined by the 2-ΔΔCt method (Livak and Schmittgen, 2001). All data were shown as mean±SD. Statistical analysis was performed using IBM SPSS 20. The data obtained from qRT-PCR analysis were subjected to One-way Analysis of Variance (ANOVA) followed by Dunnett's 2-sided post-hoc test. Significance was set at P < 0.05.
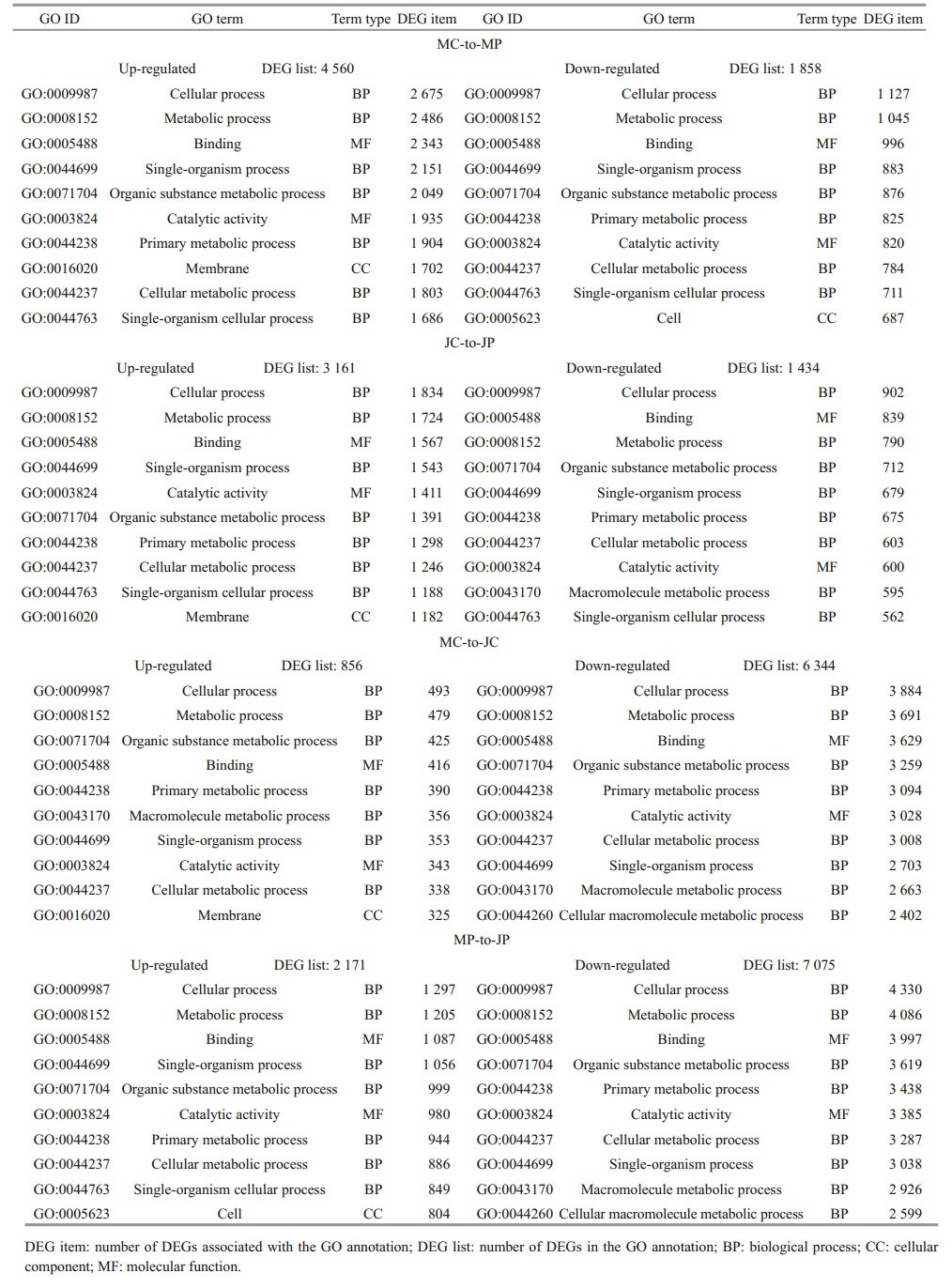
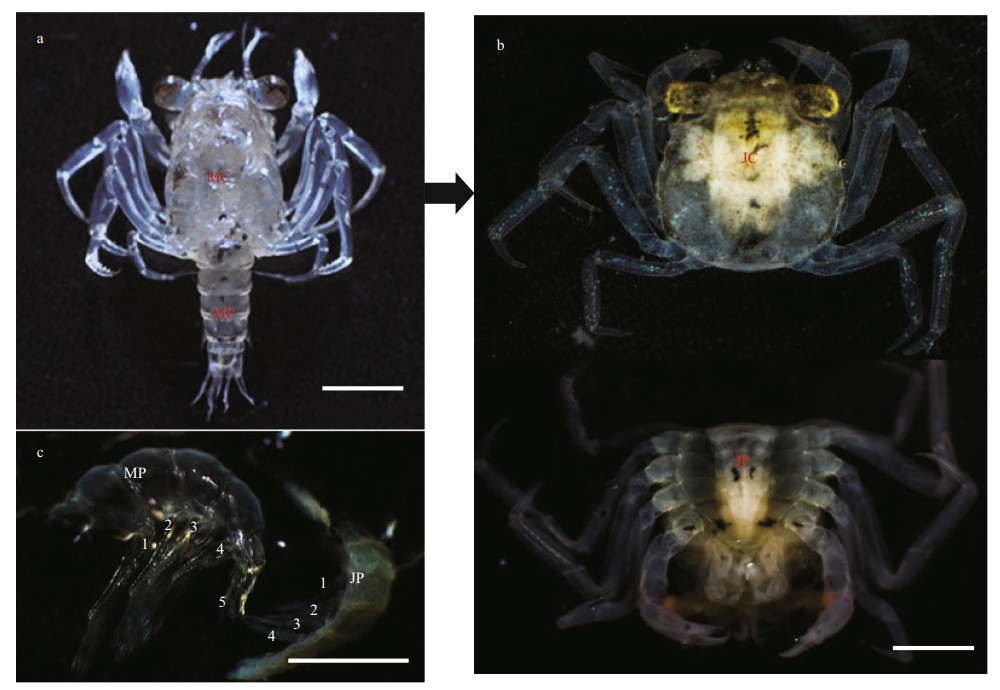
3 RESULT 3.1 External characteristics of pleonsAfter the M molted to become the J stage, the pleon was shorter, compressed, and folded beneath the cephalothorax by morphometry. The fifth pair of pleopods was smaller in M and completely reduced in J stage, and only four pairs of pleopods were observed in the pleon (Fig. 1). However, there are no external sexual characteristics between male and female crabs, in M and J stage.
|
| Fig.1 Megalopaes and the first juvenile crab stage of E. sinensis The cephalothorax of megalopaes (MC) and pleon of megalopaes (MP) (a); and the cephalothorax of the first juvenile crab stage (JC) and pleon of the first juvenile crab stage (JP) (b) were sampled for transcriptomic expression analysis, respectively. The number of pleopods in the pleon of megalopaes and the stage 1 juvenile (c). Scale bars=1 mm. |
To cover the transcriptome of cephalothoraxes and pleons in megalopaes and the first juvenile crab stage (Fig. 1), 12 cDNA libraries (three biological replicates) of E. sinensis were sequenced using Illumina Hiseq 2500 platform. The average raw reads for MC, MP, JC, and JP were 55 921 961, 52 237 985, 45 869 354, and 47 598 691, respectively. The cycleQ30 percentage of all samples was more than 91.69% (Table 1). A total of 267 158 transcripts were assembled, with a mean length of 1 123 bp using Trinity and Corset software. From these transcripts, 131 138 unigenes were assembled, with an average length of 958 bp (Supplementary Fig.S1).
Seven different databases, including Nr, Nt, KEGG, SwissProt, Pfam, GO, and KOG were used to annotate the function of 131 138 unigenes. Among the unigenes, 63 303 were annotated with at least one database, and 2 592 were annotated in all seven databases. A summary of functional annotation of contigs is showed in Supplementary Table S2.
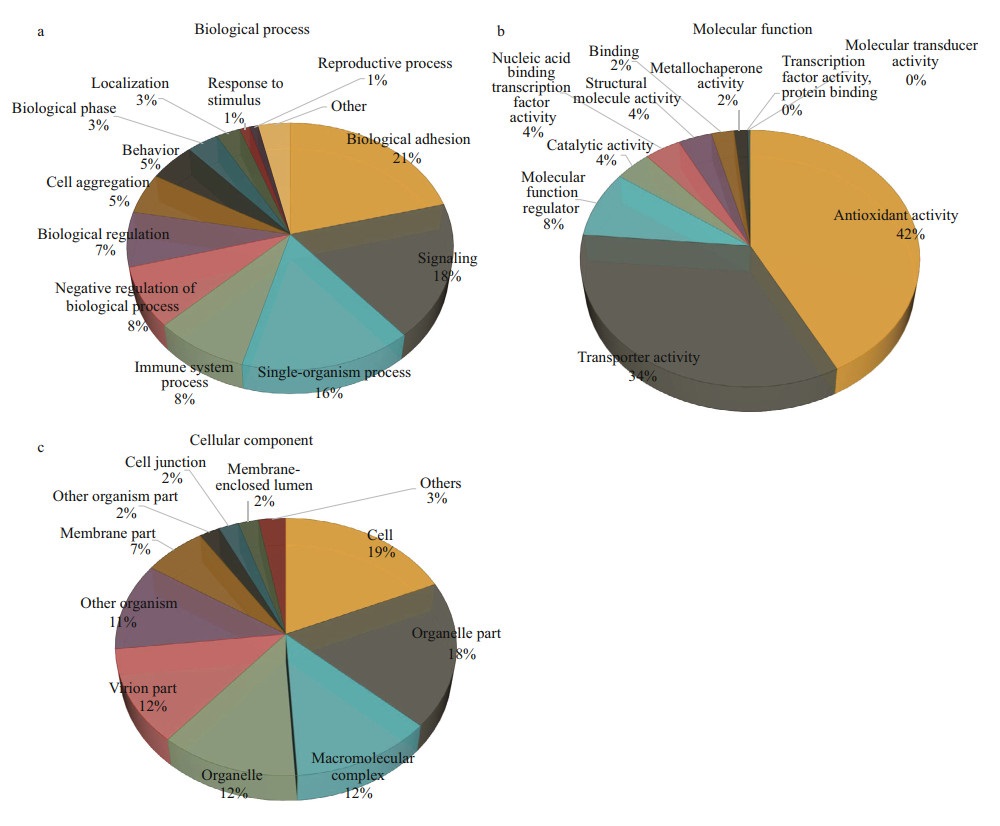
GO enrichment analysis (Supplementary Fig.S2) showed that 45 736 unigenes were associated with 56 GO terms, 26 of which were involved in biological processes (35 167 unigenes), 10 in molecular functions (36 721 unigenes) and 20 in cellular components (26 892 unigenes). The most prominent terms for biological processes, molecular functions, and cellular components are presented in Fig. 2. KOG analysis showed that 15 974 unigenes were assigned to 25 KOG terms. The top four KOG terms were general function prediction (2 156 unigenes); translation, ribosomal structure and biogenesis (2 138 unigenes); posttranslational modification, protein turnover, chaperones (1 944 unigenes); and signal transduction mechanisms (1 762 unigenes) (Supplementary Fig.S3). KEGG analysis revealed that 7 658 unigenes assigned to 5 categories which were comprised of 229 KEGG pathways. The most prominent KEGG pathways were related to ribosome (610 unigenes), carbon metabolism (234 unigenes), phagosome (193 unigenes), oxidative phosphorylation (172 unigenes), purine metabolism (157 unigenes), protein processing in endoplasmic reticulum (153 unigenes) and biosynthesis of amino acids (152 unigenes). Based on the Pfam database, 1 040 unigenes were assigned to TFs, and the main TF families were zf-C2H2 (442 unigenes), ZBTB (113 unigenes), Homeobox (79 unigenes), and bHLH (66 unigenes).
|
| Fig.2 Most prominent GO terms assigned to the transcriptome of E. sinensis The most prominent GO terms assigned to different levels of biological processes (a), molecular function (b) and cellular component (c) are listed, as calculated by Blast2GO software suite. |
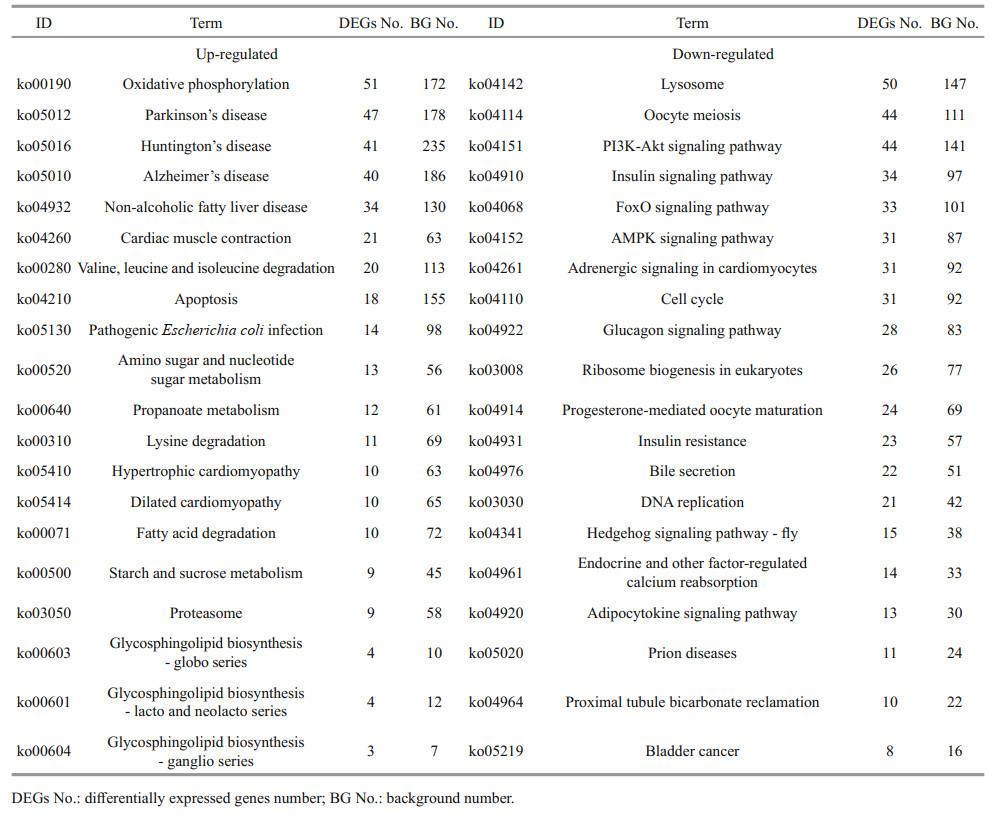
The analysis of four transcriptome data revealed 27 023 DEGs. The comparative analysis identified 9 309 DEGs (5 538 up-regulated and 3 771 downregulated) between MC and MP, 9 090 (6 650 upregulated and 2 440 down-regulated) between JC and JP, 12 760 (1 670 up-regulated and 11 090 downregulated) between MC and JC, and 16 907 (4 564 up-regulated and 12 343 down-regulated) between MP and JP. Some DEGs from four pairwise comparisons are listed in Supplementary Table S3. The distribution of DEGs was illustrated in volcano plots (Supplementary Fig.S4). Based on the GO analysis, the DEGs were mainly assigned to three functional groups: biological process, cellular component, and molecular function (Supplementary Fig.S5). In each comparison, top 10 GO terms of the most up-regulated and down-regulated DEGs are listed in Table 2. The most prominent terms for biological processes were cellular process and metabolic process; for molecular functions were binding and catalytic activity; for cellular components were membrane and cell. The top 20 KEGG pathway of the four pairwise comparisons are shown in Supplementary Fig.S6.
A homeobox TF and a fork-head TF, which are related to axial patterning, were identified among 91 dramatically up-regulated TFs during carcinization in JP compared with MP (Supplementary Table S4). In addition, a number of multifunctional TFs were also up-regulated, including the E26 transforming-specific protein (ETS), AT-rich interaction domain (ARID), E2F TFs, MYB TFs, P53 TFs, the basic leucine zipper protein of TF family (TF-bZIP), the thanatosassociated protein (THAP), basic helix-loophelix protein (bHLH) and LPS-induced tumor necrosis factor alpha factor (zf-LITAF-like). They regulate biological processes such as cell cycle, cellular differentiation, cell proliferation and apoptosis. Additionally, a Doublesex/MAB-3 domain (DM) was significantly up-regulated in JP.
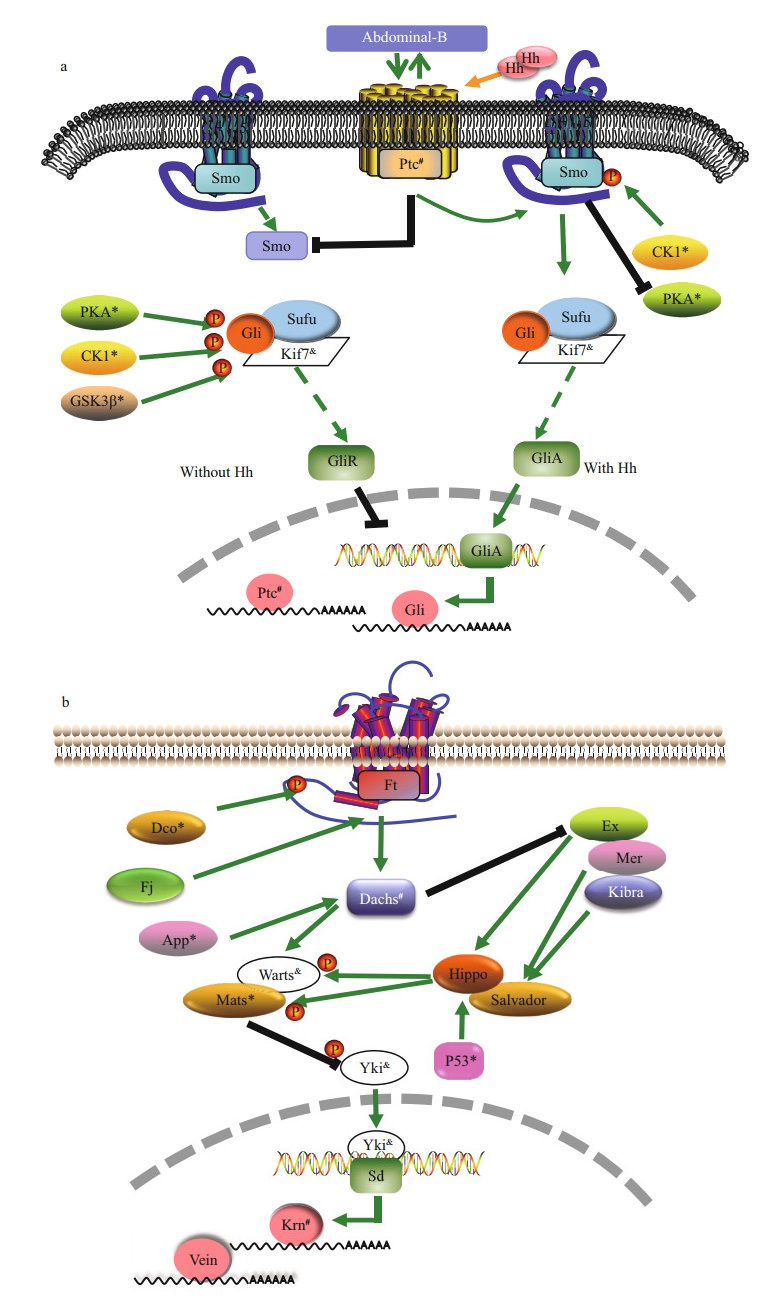
The most up-regulated top 20 KEGG pathways in MP-to-JP comparison are listed in Table 3. Regarding the energy metabolism pathways, unigenes involved in 5′-AMP-activated protein kinase (AMPK) signaling pathway were up-regulated (Fig. 3a). In the Hedgehog pathway (morphogenesis pathway), PKA, casein kinase 1 (CK1) and glycogen synthase kinase 3 beta (GSK3β) were up-regulated in JP (Fig. 4a). Meanwhile, in the Hippo pathway, discs overgrown (Dco), approximated (App), MOB kinase activator 1 (Mats) and p53 were up-regulated in JP (Fig. 4b).

|
| Fig.3 Putative energy metabolism pathways in MP-to-JP comparisons a. AMPK signaling pathway; b: oxidative phosphorylation pathway. Pathways are constructed based on KEGG pathway. The green denotes up-regulated genes in pleon of the first juvenile crab stage (JP), the down-regulated genes are labeled with purple, and yellow denotes ambiguous expression between pleon of megalopaes and JP. ND: NADH-ubiquinone oxidoreductase chain; Ndufs: NADH dehydrogenase (ubiquinone) Fe-S protein; Ndufv: NADH dehydrogenase (ubiquinone) flavoprotein; Ndufa: NADH dehydrogenase (ubiquinone) 1 alpha subcomplex subunit; Ndufb: NADH dehydrogenase (ubiquinone) 1 beta subcomplex subunit; SDHB: succinate dehydrogenase (ubiquinone) iron-sulfur subunit; ISP: ubiquinol-cytochrome c reductase iron-sulfur subunit; QCR: ubiquinol-cytochrome c reductase subunit; COX: cytochrome c oxidase subunit; OSCP: F-type H+-transporting ATPase subunit O; AMPK: 5′-AMP-activated protein kinase; FBP: fructose-1, 6-bisphosphatase I; PEPCK: phosphoenolpyruvate carboxykinase; PP2A: serine/threonine-protein phosphatase 2A catalytic subunit; TBC1D1: TBC1 domain family member 1; HSL: hormone-sensitive lipase; SCD1: stearoyl-CoA desaturase (Delta-9 desaturase); MCD: malonylCoA decarboxylase; PDK1/2: 3-phosphoinositide dependent protein kinase-1; S6K: ribosomal protein S6 kinase beta. Accessed on 2020-1-21 from https://www.genome.jp/kegg-bin/show_pathway?hsa00190. Copyright 2020, Kanehisa Laboratories. |
|
| Fig.4 Putative morphological change pathways in MP-to-JP comparison a. hedgehog signaling pathway; b. hippo signaling pathway-fly. Pathways are constructed based on KEGG pathway. *: the up-regulated genes in pleon of the first juvenile crab stage (JP); #: down-regulated genes; & : unfound genes. Hh: hedgehog; CK1: casein kinase 1; PKA: protein kinase A; Sufu: suppressor of fused; GSK3β: glycogen synthase kinase 3 beta; Ptc: patched; GliR: Gli repressor form; GliA: Gli active form; Ft: protocadherin Fat; Dco: discs overgrown; App: approximated; Mats: MOB kinase activator 1; Sd: scalloped; Krn: keren. |
The most down-regulated top 20 KEGG pathways in MP-to-JP comparison are also listed in Table 3. Unigenes involved in another energy metabolism pathways oxidative phosphorylation pathway were dramatically down-regulated (Fig. 3b). In the Hedgehog pathway, patched (Ptc) was down-regulated in JP (Fig. 4a). As to the Hippo pathway, Dachs and keren (Krn), were down-regulated in JP (Fig. 4b). Unigenes involved in apoptosis related pathways, including parkinson's disease, huntington's disease, alzheimer's disease in humans and apoptosis pathways were dramatically down-regulated in JP compared with MP (Fig. 5).

|
| Fig.5 Putative the apoptosis and apoptosis-related pathways in MP-to-JP comparisons Green line: apoptosis pathway; black line: the Parkinson's disease pathway; purple line: the Huntington's disease pathway; pink line: the Alzheimer's disease pathway; red line: common pathway. Pathways are constructed based on KEGG pathway. *: the up-regulated genes in pleon of megalopaes (MP); #: down-regulated genes; & : unfound genes; *#: ambiguous expression genes. TNF-R1: tumor necrosis factor receptor superfamily member 1A; FADD: FASassociated death domain protein; TRADD: tumor necrosis factor receptor type 1-associated DEATH domain protein; PARP: poly [ADP-ribose] polymerase 2/3/4; DAT: dopamine transporter; Cx: cytochrome c oxidase subunit; NMDAR: N-methyl-D-aspartate receptor; PSD95: postsynaptic density-95; ABAD: amyloid-binding alcohol dehydrogenase. |
Analysis of the DEGs revealed that abd-A was higher in MP (P < 0.01) than in MC, and abd-B was higher in JP (P < 0.01) than in JC (Supplementary Table S3).
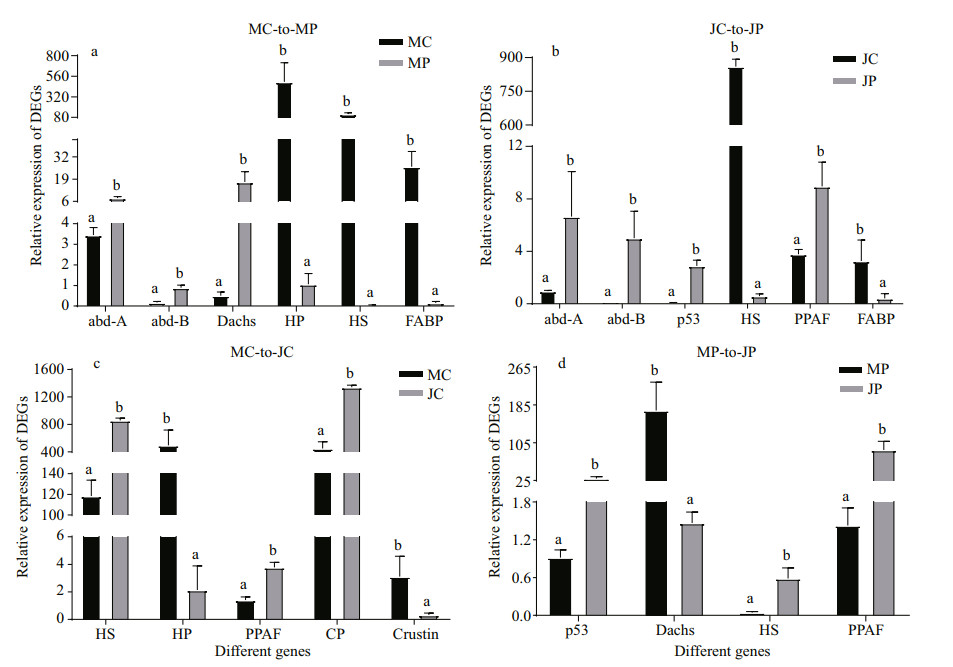
3.8 Validation of Illumina sequencing results by qRT-PCRThe expression levels of key DEGs in Hedgehog and Hippo pathways and other selected genes shown in qRT-PCR showed a high consistency with the results of RNA-Seq (Fig. 6).
|
| Fig.6 qRT-PCR validation of the selected DEGs in RNA-Seq a. MC-to-MP comparison; b. JC-to-JP comparison; c. MC-to-JC comparison; d. MP-to JP comparison. Each bars represents the mean of triplicate assays within ±SD. MC: cephalothorax of megalopaes; MP: pleon of megalopaes; JC: cephalothorax of the first juvenile crab stage; JP: pleon of the first juvenile crab stage; abd-A: abdominal-A; abd-B: abdominal-B; HP: hypothetical protein T11_4388; HS: hemocyanin subunit 6; FABP: fatty acid binding protein; PPAF: prophenoloxidase-activating factor; CP: cuticle proprotein proCP2.2. |
Crabs undergo morphological changes after molting to the first juvenile crab stage. Compared to the megalopae with five pairs of pleopos (Montú et al., 1996), the pleopods were degenerate, only four pairs of pleopods in J stage. The development of pleopods in E. sinensis is similar to that in the Japanese mitten crab, Eriocheir japonicus (Lee et al., 1994) and the mud crab Scylla paramamosain (Jiang et al., 2020). High-throughput sequencing is a newly developed technology for discovering and profiling genes efficiently (Reuter et al., 2015). A previous study used the suppression subtractive hybridization (SSH) method to indetify the DEGs during the carcinization of the Chinese mitten crab assembled only 325 unigenes (Li et al., 2011), but with highthroughput sequencing, 16 907 different expression genes were assembled between MP and JP. From a comparison with SSH, the RNA-Seq generated more data. The most prominent KEGG pathways in the development of M to J stage (MC-to-JC and MP-toJP) of E. sinensis were similar to the metamorphosis research in the prawn Macrobrachium rosenbergii (Ventura et al., 2013), which also undergoes morphological change from planktonic larval stages to the benthic post-larval stage (Abdu et al., 1998).
4.1 Energy metabolism during carcinizationIn the oxidative phosphorylation pathway, NADH dehydrogenase, cytochrome c reductase, cytochrome c oxidase and F-type ATPase work together to generate energy (Dimroth et al., 1999; Heikal et al., 2014). The AMPK pathway is a canonical route regulating energy homeostasis of the whole body (López et al., 2016). In this study, the oxidative phosphorylation pathway had the most downregulated genes, while genes related to AMPK signaling pathway were up-regulated, in MP compared with JP. This result suggests that the energy metabolism pathway have changed in the pleon from the M to J stage, the main pathway was the AMPK pathway rather than the oxidative phosphorylation pathway in J stage, and the mechanism of energy utilization have changed. The pleon of megalopae crabs remains extended for better swimming ability (Atkins, 1954), while the pleon of the juvenile mitten crab is tucked under the cephalothorax in an adaptation to a benthic lifestyle, now protecting reproductive organs for mating and carrying eggs in adult female crabs (Števčić, 1971; Herborg et al., 2006; Davie et al., 2015). It is likely that the changed energy metabolism pathways of the pleon coincide with the adjustment of swimming ability, as describe in the bryozoan Bugula neritina (Wong et al., 2014), in which the level of energy production decreased, when the larvae ceased swimming.
4.2 Morphological changes of pleon in J stageTFs have a leading role in the initiation of gene expression (Spitz and Furlong, 2012). In this study, 91 TFs were dramatically up-regulated in JP compared with MP, among which 13 zf-C2H2 TFs were identified. The Homeobox TF and fork-head TF play an important role in the development of organisms, such as Strongylocentrotus purpuratus (HowardAshby et al., 2006) and Xenopus laevis (Rohl and Knöchel, 2005). As a class of zinc finger motifs, zfC2H2 TFs execute important regulatory functions in the development of Strongylocentrotus purpuratus (Materna et al., 2006). Therefore, we deduced that zfC2H2 TFs as well as Homeobox TF and fork-head TF may be involved in the pleon development from M to J stage expecially in the morphological change by regulating target gene expression. Futhermore, a DM was also up-regulated in MP-to-JP comparisons. DM domain has been shown to be involved in sex-specific differentiation of Caenorhabditis elegans and the medaka, Oryzias latipes (Lints and Emmons, 2002; Nagahama, 2005). In the brachyuran decapods, the differentiation of gonopores and pleopods take place simultaneously from the first to the third juvenile crab stage (Lee et al., 1994). Therefore, we propose that the up-regulation of DM domain might correlate to the differentiation of gonopores or pleopods in the early juveniles.
Hedgehog signaling pathway and Hippo signaling pathway-fly that up-regulated in JP compared with MP, are implicated in the regulation of metamorphosis of many metazoan groups (Ingham and McMahon, 2001; Pan, 2007). The Hedgehog pathway is involved in organizing growth and patterning in many developmental processes (Mohler, 1988) and the Hippo pathway coordinates with cell death, differentiation and inhibition of cell proliferation (Yu and Guan, 2013). Our transcriptomic analysis showed that the DEGs in MP-to-JP comparison were assigned to Hedgehog pathway, which was similar to the previous study of the carcinization of E. sinensis (Song et al., 2015). Additionally, we found the Hippo pathway might take part in the carcinization of the Chinese mitten crab in this research.
The Hox gene abd-B plays an important role in appendage development. For example, it suppresses the development of lepidopteran proleg in the posterior abdomen and suppresses the development of appendages in genus of Drosophila (Estrada and Sánchez-Herrero, 2001; Tomita and Kikuchi, 2009). Additionally, the interaction between abd-B and Hedgehog pathway regulates the chick hindgut and Drosophila genital disc development (Roberts et al., 1995; Sánchez et al., 2001). Analysis of the DEGs showed that the abd-B was higher in JP (P < 0.01) than in JC. Hence, we deduced that abd-B might participate in the degeneration of the fifth pair of pleopods during carcinization by regulating the Hedgehog signaling pathway.
4.3 The fusion of ventral nerve cord during carcinizationApoptosis or programmed cell death (PCD) is a conserved process among species (Vaux et al., 1994). We found that the unigenes involved in apoptosis related pathways were dramatically down-regulated in JP compared with MP. In carcinization, the ventral nerve cord fuses with the thoracic ganglion to form the thoracic ganglion in the thorax (Davie et al., 2015). In the Chinese mitten crab, E. sinensis, the fusion of ventral nerve cord and thoracic ganglion begins in M stage and completes in J stage (Song et al., 2017). Apoptosis-related pathway including, parkinson's disease, huntington's disease, and alzheimer's disease in humans, as well as apoptosis pathway are related to central nervous system development and neurodegeneration (Schmidt et al., 1997; Federico et al., 2012). Furthermore, abdominal neuroblasts stop dividing at mid-larval stages due to the activation of apoptosis, which is mediated by the Hox gene abd-A, in Drosophila (Bello et al., 2003). Analysis of the transcriptome data, we found that the abd-A was higher in MP (P < 0.01) than in MC, which suggested the abd-A might involve in the fusion of ventral nerve cord during carcinization.
5 CONCLUSIONIn this study, we reported the transcriptomic changes during megalopa molting to the first juvenile crab stage of E. sinensis. Our results reveal major transcriptomic changes of pleons in terms of the oxidative phosphorylation and the AMPK pathways of the energy metabolism, the TFs, the Hedgehog and Hippo pathway of the morphological changes, and apoptosis-related pathways during the fusion of ventral nerve cord. By studying the key Hox genes abd-B and abd-A in carcinization, we found that the abd-B might participate in the degeneration of pleopod by regulating the Hedgehog signalling pathway-fly and the abd-A might regulate the ganglion fusion.
6 DATA AVAILABILITY STATEMENTAll the original Illumina HiSeq sequencing data are available from the NCBI Sequence Read Archive (SRA) database (PRJNA644959). Other data that support the current study are available from the corresponding author on reasonable request.
Electronic supplementary materialSupplementary material (Supplementary Tables S1–S4 and Figs.S1–S6) is available in the online version of this article at https://doi.org/10.1007/s00343-020-0176-5.
Abdu U, Takac P, Laufer H, Sagi A. 1998. Effect of methyl farnesoate on late larval development and metamorphosis in the prawn Macrobrachium rosenbergii (Decapoda, Palaemonidae): a juvenoid-like effect?. The Biological Bulletin, 195(2): 112-119.
DOI:10.2307/1542818 |
Anger K. 1991. Effects of temperature and salinity on the larval development of the Chinese mitten crab Eriocheir sinensis (Decapoda: grapsidae). Marine Ecology Progress Series, 72: 103-110.
DOI:10.3354/meps072103 |
Atkins D. 1954. Leg disposition in the brachyuran megalopa when swimming. Journal of the Marine Biological Association of the United Kingdom, 33(3): 627-636.
DOI:10.1017/S0025315400026904 |
Bello B C, Hirth F, Gould A P. 2003. A pulse of the Drosophila Hox protein Abdominal-A schedules the end of neural proliferation via neuroblast apoptosis. Neuron, 37(2): 209-219.
DOI:10.1016/S0896-6273(02)01181-9 |
Davie P J F, Guinot D, Ng P K L. 2015. Anatomy and functional morphology of Brachyura. In: Castro P, Davie P, Guinot D, Schram F, Von Vaupel Klein C eds. Treatise on Zoology-Anatomy, Taxonomy, Biology. The Crustacea, Volume 9 Part C (2 vols). Brill, p. 11–163.
|
Dimroth P, Wang H Y, Grabe M, Oster G. 1999. Energy transduction in the sodium F-ATPase of Propionigenium modestum. Proceedings of the National Academy of Sciences of the United States of America, 96(9): 4924-4929.
DOI:10.1073/pnas.96.9.4924 |
Estrada B, Sánchez-Herrero E. 2001. The Hox gene Abdominal-B antagonizes appendage development in the genital disc of Drosophila. Development, 128(3): 331-339.
DOI:10.1242/dev.128.3.331 |
FAO. 2020. Food and Agriculture Organization of the United Nations Global Production Statistics. http://www.fao.org/fishery/statistics/global-aquaculture-production/query/en. Accessed on 2020-04-16.
|
Federico A, Cardaioli E, Da Pozzo P, Formichi P, Gallus G N, Radi E. 2012. Mitochondria, oxidative stress and neurodegeneration. Journal of the Neurological Sciences, 322(1-2): 254-262.
DOI:10.1016/j.jns.2012.05.030 |
Garber M, Grabherr M G, Guttman M, Trapnell C. 2011. Computational methods for transcriptome annotation and quantification using RNA-seq. Nature Methods, 8(6): 469-477.
DOI:10.1038/nmeth.1613 |
Guinot D, Bouchard J M. 1998. Evolution of the abdominal holding systems of brachyuran crabs (Crustacea, Decapoda, Brachyura). Zoosystema, 20(4): 613-694.
|
Heikal A, Nakatani Y, Dunn E, Weimar M R, Day C L, Baker E N, Lott J S, Sazanov L A, Cook G M. 2014. Structure of the bacterial type II NADH dehydrogenase: a monotopic membrane protein with an essential role in energy generation. Molecular Microbiology, 91(5): 950-964.
DOI:10.1111/mmi.12507 |
Herborg L M, Bentley M G, Clare A S, Last K S. 2006. Mating behaviour and chemical communication in the invasive Chinese mitten crab Eriocheir sinensis. Journal of Experimental Marine Biology and Ecology, 329(1): 1-10.
DOI:10.1016/j.jembe.2005.08.001 |
Howard-Ashby M, Materna S C, Brown C T, Chen L L, Cameron R A, Davidson E H. 2006. Identification and characterization of homeobox transcription factor genes in Strongylocentrotus purpuratus, and their expression in embryonic development. Developmental Biology, 300(1): 74-89.
DOI:10.1016/j.ydbio.2006.08.039 |
Ingham P W, McMahon A P. 2001. Hedgehog signaling in animal development: paradigms and principles. Genes & Development, 15(23): 3059-3087.
|
Jiang Q L, Lu B, Lin D D, Huang H Y, Chen X L, Ye H H. 2020. Role of crustacean female sex hormone (CFSH) in sex differentiation in early juvenile mud crabs, Scylla paramamosain. Generaland Comparative Endocrinology, 289: 113383.
DOI:10.1016/j.ygcen.2019.113383 |
Langmead B. 2010. Aligning short sequencing reads with Bowtie. Current Protocols in Bioinformatics, 32(1): 11.7.1-11.7.14.
|
Lee T H, Yamauchi M, Yamazaki F. 1994. Sex differentiation in the crab Eriocheir japonicus (Decapoda, Grapsidae). Invertebrate Reproduction & Development, 25(2): 123-137.
|
Li B, Dewey C N. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics, 12: 323.
DOI:10.1186/1471-2105-12-323 |
Li P, Zha J, Sun H Y, Song D X, Zhou K Y. 2011. Identification of differentially expressed genes during the brachyurization of the Chinese mitten crab Eriocheir japonica sinensis. Biochemical Genetics, 49(9-10): 645-655.
DOI:10.1007/s10528-011-9439-3 |
Lints R, Emmons S W. 2002. Regulation of sex-specific differentiation and mating behavior in C.elegans by a new member of the DM domain transcription factor family.. Genes & Development, 16(18): 2390-2402.
|
Livak K J, Schmittgen T D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods, 25(4): 402-408.
DOI:10.1006/meth.2001.1262 |
López M, Nogueiras R, Tena-Sempere M, Diéguez C. 2016. Hypothalamic AMPK: a canonical regulator of whole-body energy balance. Nature Reviews Endocrinology, 12(7): 421-432.
DOI:10.1038/nrendo.2016.67 |
Love M I, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology, 15(12): 550.
DOI:10.1186/s13059-014-0550-8 |
Materna S C, Howard-Ashby M, Gray R F, Davidson E H. 2006. The C2H2 zinc finger genes of Strongylocentrotus purpuratus and their expression in embryonic development. Developmental Biology, 300(1): 108-120.
DOI:10.1016/j.ydbio.2006.08.032 |
McLaughlin P A, Lemaitre R. 1997. Carcinization in the Anomura-fact or fiction? I.Evidence from adult morphology. Contributions to Zoology, 67(2): 79-123.
DOI:10.1163/18759866-06702001 |
Medina M. 2009. Functional genomics opens doors to understanding metamorphosis in nonmodel invertebrate organisms. Molecular Ecology, 18(5): 763-764.
DOI:10.1111/j.1365-294X.2008.04079.x |
Mohler J. 1988. Requirements for hedgehog, a segmental polarity gene, in patterning larval and adult cuticle of Drosophila. Genetics, 120(4): 1061-1072.
DOI:10.1093/genetics/120.4.1061 |
Montú M, Anger K, De Bakker C. 1996. Larval development of the Chinese mitten crab Eriocheir sinensis H.Milne-Edwards (Decapoda: Grapsidae) reared in the laboratory. Helgoländer Meeresuntersuchungen, 50(2): 223-252.
DOI:10.1007/BF02367153 |
Nagahama Y. 2005. Molecular mechanisms of sex determination and gonadal sex differentiation in fish. Fish Physiology and Biochemistry, 31(2-3): 105.
DOI:10.1007/s10695-006-7590-2 |
Pan D J. 2007. Hippo signaling in organ size control. Genes & Development, 21(8): 886-897.
|
Payen G G. 1975. Effects masculinisants des glandes androgenes implantees chez la femelle pubere pedonculec'tomisee de Rhizocephalan harrisii (Gould) (Crustace, Decapode, Brachyoure). Comptes Rendus des Seances de l'Academie des Sciences, Serie D, 280: 1111-1114.
|
Reuter J A, Spacek D V, Snyder M P. 2015. High-throughput sequencing technologies. Molecular Cell, 58(4): 586-597.
DOI:10.1016/j.molcel.2015.05.004 |
Roberts D J, Johnson R L, Burke A C, Nelson C E, Morgan B A, Tabin C. 1995. Sonic hedgehog is an endodermal signal inducing Bmp-4 and Hox genes during induction and regionalization of the chick hindgut. Development, 121(10): 3163-3174.
DOI:10.1242/dev.121.10.3163 |
Rohl B S, Knöchel W. 2005. Of fox and frogs: fox (fork head/winged helix) transcription factors in Xenopus development. Gene, 344: 21-32.
DOI:10.1016/j.gene.2004.09.037 |
Sánchez L, Gorfinkiel N, Guerrero I. 2001. Sex determination genes control the development of the Drosophila genital disc, modulating the response to Hedgehog, Wingless and Decapentaplegic signals. Development, 128(7): 1033-1043.
DOI:10.1242/dev.128.7.1033 |
Schmidt H, Rickert C, Bossing T, Vef O, Urban J, Technau G M. 1997. The embryonic central nervous system lineages of Drosophila melanogaster. Developmental Biology, 189(2): 186-204.
DOI:10.1006/dbio.1997.8660 |
Scholtz G. 2014. Evolution of crabs—history and deconstruction of a prime example of convergence. Contributions to Zoology, 83(2): 87-105.
DOI:10.1163/18759866-08302001 |
Song C W, Cui Z X, Hui M, Liu Y, Li Y D, Li X H. 2015. Comparative transcriptomic analysis provides insights into the molecular basis of brachyurization and adaptation to benthic lifestyle in Eriocheir sinensis. Gene, 558(1): 88-98.
DOI:10.1016/j.gene.2014.12.048 |
Song D L, Li Y D, Cao Y C, Zhang H Y, Guo E M. 2017. Histological observation of nervous system and postembryonic development of Chinese mitten crab Eriocheir sinensis. Fisheries Science, 36(2): 183-187.
(in Chinese with English abstract) |
Spitz F, Furlong E E M. 2012. Transcription factors: from enhancer binding to developmental control. Nature Reviews Genetics, 13(9): 613-626.
DOI:10.1038/nrg3207 |
Števčić Z. 1971. The main features of brachyuran evolution. Systematic Biology, 20(3): 331-340.
|
Sui L Y, Wille M, Cheng Y X, Wu X G, Sorgeloos P. 2011. Larviculture techniques of Chinese mitten crab Eriocheir sinensis. Aquaculture, 315(1-2): 16-19.
DOI:10.1016/j.aquaculture.2010.06.021 |
Tavazoie S, Hughes J D, Campbell M J, Cho R J, Church G M. 1999. Systematic determination of genetic network architecture. Nature Genetics, 22(3): 281-285.
DOI:10.1038/10343 |
Tomita S, Kikuchi A. 2009. Abd-B suppresses lepidopteran proleg development in posterior abdomen. Developmental Biology, 328(2): 403-409.
DOI:10.1016/j.ydbio.2009.01.040 |
Trapnell C, Williams B A, Pertea G, Mortazavi A, Kwan G, Van Baren M J, Salzberg S L, Word B J, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology, 28(5): 511-515.
DOI:10.1038/nbt.1621 |
Vaux D L, Haecker G, Strasser A. 1994. An evolutionary perspective on apoptosis. Cell, 76(5): 777-779.
DOI:10.1016/0092-8674(94)90350-6 |
Ventura T, Manor R, Aflalo E D, Chalifa-Caspi V, Weil S, Sharabi O, Sagi A. 2013. Post-embryonic transcriptomes of the prawn Macrobrachium rosenbergii: multigenic succession through metamorphosis. PLoS One, 8(1): e55322.
DOI:10.1371/journal.pone.0055322 |
Wong Y H, Ryu T, Seridi L, Ghosheh Y, Bougouffa S, Qian P Y, Ravasi T. 2014. Transcriptome analysis elucidates key developmental components of bryozoan lophophore development. Scientific Reports, 4: 6534.
|
Yu F X, Guan K L. 2013. The Hippo pathway: regulators and regulations. Genes & Development, 27(4): 355-371.
|
 2021, Vol. 39
2021, Vol. 39