Institute of Oceanology, Chinese Academy of Sciences
Article Information
- LIU Wenzheng, LIU Qianchun, ZHAO Jin, WEI Xiu, JIANG Peng
- Comparative chloroplast genomes of Ulva prolifera and U. linza (Ulvophyceae) provide genetic resources for the development of interspecific markers
- Journal of Oceanology and Limnology, 40(6): 2372-2384
- http://dx.doi.org/10.1007/s00343-022-2045-x
Article History
- Received Jan. 29, 2022
- accepted in principle Apr. 2, 2022
- accepted for publication May 9, 2022
2 Laboratory for Marine Biology and Biotechnology, Pilot National Laboratory for Marine Science and Technology(Qingdao), Qingdao 266237, China;
3 University of Chinese Academy of Sciences, Beijing 100049, China;
4 College of Life Science, Qingdao University, Qingdao 266071, China
Ulva species are widely distributed worldwide, thriving in intertidal, brackish, estuaries, and even freshwater environments (Mantri et al., 2020), with more than 80 identified species documented in the Algaebase (Guiry and Guiry, 2021). The thallus of Ulva is composed of distromatic blade or monostromatic tube. As their morphological features are very limited and unstable, which are sensitive to various factors such as salinity (Blomster et al., 1998), temperature (Blomster et al., 2002), and associated bacteria (Kessler et al., 2018), the morphological identification for Ulva are always very difficult (Blomster et al., 1999). The development of molecular approaches has significantly improved this dilemma, resulting in the reconstruction of genera Ulva (Hayden et al., 2003), and identification of some cryptic species (Hofmann et al., 2010), but some of related species still lack appropriate molecular markers to distinguish them from each other (Kang et al., 2019; Steinhagen et al., 2019).
The type locations for U. linza Linnaeus 1753 and U. prolifera O. F. Müller 1778 were at Kent of England, and Danish island of Lorand respectively. Due to the lack of holotype of U. prolifera, the representative sequences of genetic markers for this species were originally derived from the samples collected in the British Isles that have been identified as "U. prolifera" on the basis of morphological characteristics (Blomster et al., 1998; Tan et al., 1999). However, Shimada et al. (2008) suggested that in the ITS-based phylogenetic tree, those U. prolifera collected in Japan were separated from European "U. prolifera", and they were almost completely indistinguishable with U. linza and U. procera (synonym of U. prolifera), forming a cluster named LPP. Cui et al. (2018) confirmed that the epitype of U. prolifera collected from the type location were also located in the LPP cluster, and suggested to revise the previous "U. prolifera" to U. splitiana. In addition to the genetic similarities between U. prolifera and U. linza, their distribution areas are often overlapped as well, including the Baltic Sea in Europe (Cui et al., 2018; Steinhagen et al., 2019), the Atlantic coast of North America (Guidone et al., 2013), and many parts of the Northwest Pacific (Shimada et al., 2008; Zhao et al., 2018). In general, U. linza is mainly spread in marine habitats, while strains of U. prolifera are found commonly in estuaries and brackish waters (Shimada et al., 2008; Ogawa et al., 2013). However, in the Southern Yellow Sea area, the two species grew intermixed and the biomass of both species is very high (Han et al., 2013). In particular, U. prolifera, as the dominant species, has caused the largest green tide in the world for consecutive years (Zhao et al., 2013). Thus, the accurate discrimination between these two related species have become necessary for investigations of Ulva species composition and green tide monitoring especially in this sea area.
Because molecular markers commonly used in Ulva, including ITS, rbcL and tufA, always failed to distinguish between U. prolifera and U. linza (Leliaert et al., 2009; Zhang et al., 2011; Xie et al., 2019), the 5S rDNA spacer region, which was polymorphic in individual, was developed (Shimada et al., 2008), and has been used widely to discriminate these two species (Hiraoka et al., 2011; Duan et al., 2012; Zhang et al., 2015; Song et al., 2019). From each species, this marker could generate multiple amplified products of different sequences and lengths, of which the smallest fragment of about 300 bp was considered to be specific to U. linza, and was not available in U. prolifera. However, this likely U. linza-specific genotype was later found in the epitype of U. prolifera as well (Cui et al., 2018), suggesting that the 5S rDNA spacer region was probably not a substantial interspecific marker (Melton III and Lopez-Bautista, 2021). New efforts focused more on the organelle genomes, since they have much richer polymorphic sites which are usually used for phylogenetic analysis among populations or related species (Yang et al., 2013; Zhang et al., 2021). Liu et al. (2020b) reported that a newly developed mitochondrial marker rps2-trnL can well distinguish four Ulva species including U. linza and the drifting U. prolifera causing the Yellow Sea green tide. However, the drifting U. prolifera has been revealed as a unique floating ecotype, which was clearly different from the widely-distributed attached populations, in terms of both genetics and the performances of reproductive isolation with U. linza (Hiraoka et al., 2011; Zhao et al., 2015), whether the usage of rps2-trnL can be extended to distinguish these two species still needs further verification with the attached populations of U. prolifera.
In this study, the chloroplast genome of a representative strain for attached U. prolifera was sequenced, and two existing chloroplast genomes which were from U. linza and the floating ecotype of U. prolifera respectively, were combined for a comparative analysis. The identified interspecific variations were used to develop new markers for the discrimination between these two related species.
2 MATERIAL AND METHOD 2.1 Seaweeds and molecular identificationEach Ulva strain used in this study was unialgal culture maintained in our laboratory, the collection information were shown in Supplementary Table S1. All the samples were cultured in Von Stosch's Enriched (VSE) medium renewed once a week, at 20 ℃ with a 12-h꞉12-h light (L)꞉dark (D) photoperiod and a photosynthetic irradiance of about 80 μmol photons/(m2·s).
Genomic DNA of each sample was extracted using a Plant Genomic DNA Extraction Kit (Tiangen Biotech Co. Ltd., Beijing, China) according to the manufacturer's instruction. The molecular identification for all samples were performed using ITS, 5S rDNA spacer, and a sequence characterized amplified region (SCAR) marker which was specific to the floating ecotype of U. prolifera dominating the green tide in the Yellow Sea. The primers and PCR procedures for ITS, 5S rDNA spacer, and SCAR markers referred to Leskinen and Pamilo (1997), Shimada et al. (2008), and Zhao et al. (2015) respectively. PCR products were sequenced in Ruibo Bio Tech Co. Ltd, Qingdao, China by a Genetic Analyzer (ABI3730XL, USA). Phylogenetic analysis were performed according to previous descriptions from Xie et al. (2020).
2.2 Chloroplast genome sequencing, assembly, annotation, and phylogenetic analysisAn attached U. prolifera sample U161 was selected as a representative for chloroplast genome sequencing. A single thallus was cut into segments for vegetative growth, then the algal tissue was sent to HengChuang Gene Co. Ltd. (Shenzhen, China) for high-throughput sequencing. Total genomic DNA was extracted using a Plant Genomic DNA Extraction Kit (Tiangen Biotech Co. Ltd., Beijing, China). The DNA library with an insert size of 350 bp was constructed using a library preparation kit (New England Biolabs Co. Ltd., USA) and sequenced using the Hiseq 4000 platform (Illumina Co. Ltd., USA) to obtain 150 bp ×2 paired-end reads. The low-quality sequences which are those with over 50% bases having quality values of Q < 19 or over 5% bases being 'N' were removed. The filtered reads were assembled into contigs by SOAPdenovo v2.04 (Luo et al., 2015), then aligned and ordered according to the reference genome. Last, raw reads were again mapped to the assembled draft chloroplast genome and the majority of gaps were filled through local assembly.
The chloroplast genome was annotated using program PGA (Qu et al., 2019). Ribosomal RNA genes (rRNAs) were identified by RNAmmer v1.2 (Lagesen et al., 2007), and transfer RNA genes (tRNAs) were searched using the tRNAscan-SE v2.0 (Chan and Lowe, 2019). The OGDRAW v1.3.1 was applied to draw the genome map (Greiner et al., 2019). The whole chloroplast genome sequence with annotation information was submitted to GenBank of NCBI using Bankit.
For phylogenetic analysis with whole chloroplast genomes, a total of 48 shared protein-coding genes among all available 26 chloroplast genomes of Ulva, including our data from U161 and other 25 which were obtained from NCBI as references, were selected for alignment by MAFFT v7.475 (Kuraku et al., 2013). After alignment and concatenating of the shared genes, the full length of 48 gene sequences were about 36 kb. The maximum likelihood (ML) phylogenetic tree with alignment sequences from 26 chloroplast genomes of Ulva was constructed using a GTR + G + I model and the sequence divergences were calculated with MEGA 6.0 (Tamura et al., 2013).
2.3 Comparative genomic analysis between U. linza and two ecotypes of U. proliferaThe complete chloroplast genomes of U. linza (NC030312), the floating U. prolifera (NC036137) collected from the Yellow Sea green tide, and the attached U. prolifera U161 (MZ571508), were used for comparative genomic analysis. The codon usage biases was analyzed using PhyloSiute v1.2.2 (Zhang et al., 2020) and codonW v1.4.4 (Meade et al., 1997). The collinearity analysis with these three chloroplast genomes was carried out to check the genome rearrangement by Mauve v2.4 with the ProgressiveMauve algorithm (Darling et al., 2010). Single nucleotide polymorphism (SNP) sites were searched by Mauve v2.4, and indel (insertion-deletion) sites were identified by Dnasp v5.1 (Librado and Rozas, 2009). In order to visualize structure variations across the genomes, the chloroplast genomic sequence comparative analysis were conducted using the mVISTA following a global pairwise alignment of the sequences with the LAGAN program (Frazer et al., 2004).
2.4 Development of new species-specific markers from chloroplast genomesFrom the identified SNP, indels or structural variations, some of those regions that were homologous between the two ecotypes of U. prolifera but had obvious divergences between U. prolifera and U. linza were selected as molecular marker targets, and the flanking sequences at both ends, which were completely identical among the three chloroplast genomes, were used for design of species-specific primers using Primer 3.0. All primers were synthesized by Sangon Biotech (Shanghai) Co. Ltd. (Shanghai, China). The effects of species distinguish for designed primers were evaluated with each of twelve Ulva samples by PCR reactions. The profile of the PCR reactions consisted of one initial denaturation of 10 min at 94 ℃, then 35 cycles of denaturation of 45 s at 94 ℃, primer annealing of 45 s at 55 ℃ and extensions of 2 min at 72 ℃, and a final extension of 10 min at 72 ℃. Following the cycles, there was a final hold at 4 ℃. PCR products were detected using gel electrophoresis in a 1.5% agarose gel stained with Super GelRed (US Everbright Inc., Suzhou, China). The sequencing and phylogenetic analysis were performed following the previous descriptions for ITS.
3 RESULT 3.1 Molecular identificationThe phylogenetic tree for ITS showed that all 12 samples fell into the U. prolifera - U. linza complex (Supplementary Fig.S1), and the tree for 5S rRNA spacer showed that they were clearly resolved into two clades, i.e., U. prolifera and U. linza. After that, eight samples of U. prolifera were detected by SCAR marker further, and the results showed that all four floating samples belonged to the floating ecotype of U. prolifera (Fig. 1).

|
| Fig.1 Phylogenetic tree based on ML analysis with 5S rDNA spacer sequences Numbers at the nodes indicate bootstrap values. GenBank accession numbers for all reference sequences are provided. Sequences in bold were from samples in this study. 'SCAR+' represents positive result for SCAR; 'SCAR–' represents negtive result for SCAR. |
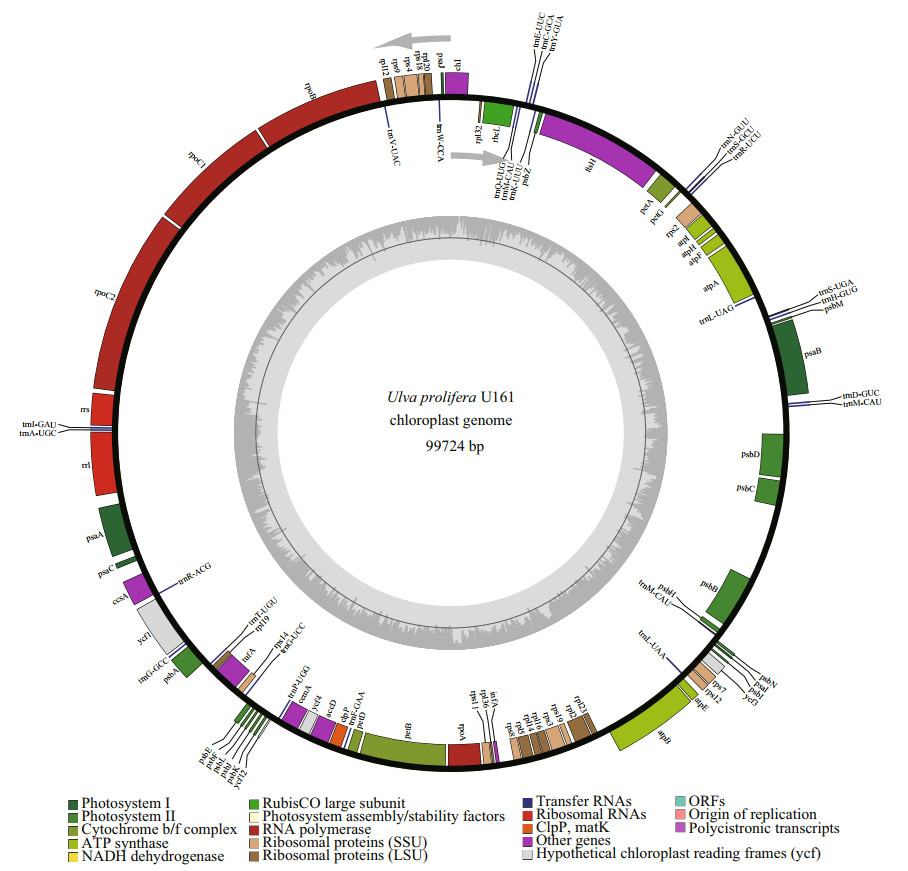
To develop interspecific genetic markers for U. prolifera and U. linza based on the intraspecific variations within U. prolifera, an attached U. prolifera strain U161 was selected for sequencing of chloroplast genome since both references of floating U. prolifera and U. linza are readily available. After genome sequencing, assembly, and annotation, it was shown that the complete chloroplast genome of U161 is 99 724 bp in size (Fig. 2) (GenBank accession No. MZ571508), encoding 95 genes including 67 protein-coding genes, 26 tRNAs, and 2 rRNAs. There are five genes (psbB, psbD, atpA, atpB, and psaB) containing one intron and there is one gene (petB) containing two introns. The overall base composition was A (37.7%), T (37.0%), C (12.6%), and G (12.7%). The voucher (assigned number MBM 287038) was deposited in the Marine Biological Museum of Chinese Academy of Sciences (MBMCAS) at the Institute of Oceanology, Chinese Academy of Sciences, China.
|
| Fig.2 Chloroplast genome map of U. prolifera U161 Genes shown inside of the circle are transcribed clockwise, while those outsides are transcribed counterclockwise. Bars of different colors belong to different functional groups. The darker gray in the inner circle corresponds to GC content, whereas the lighter gray corresponds to AT content. |
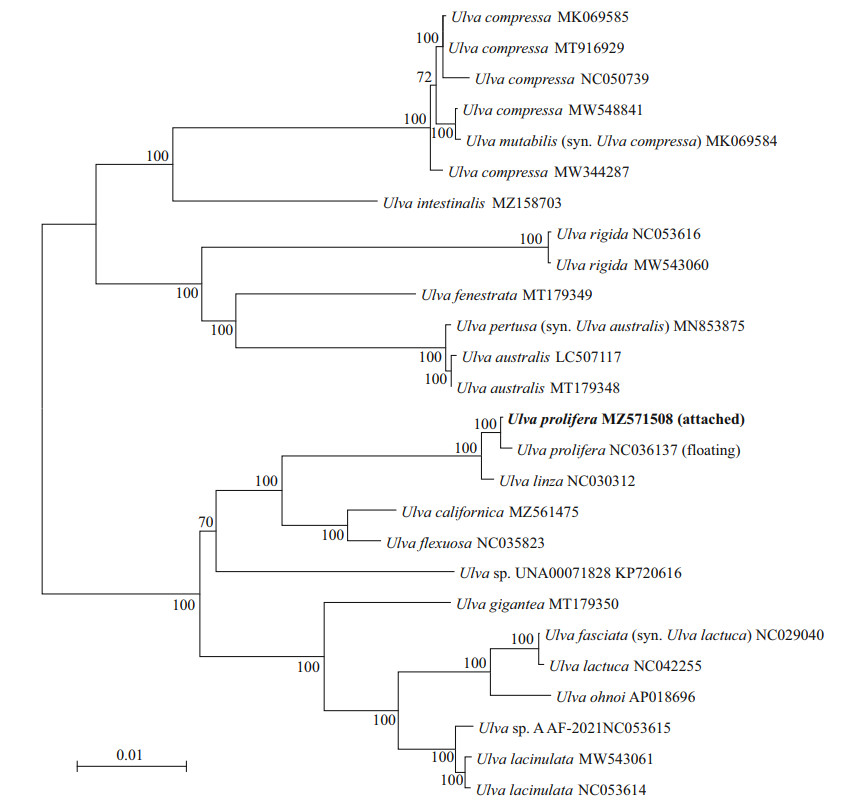
The ML phylogenetic tree of chloroplast genomes of Ulva was shown in Fig. 3. It was shown that the attached and floating U. prolifera gathered into a cluster which was separated from U. linza. The chloroplast genome sequence divergence was 0.3% between U. linza and the attached U. prolifera, and 0.4% between U. linza and the floating U. prolifera. This result suggested that U. linza and U. prolifera can be distinguished as two species by the whole chloroplast genome despite the intraspecific divergences within U. proifera.
|
| Fig.3 Phylogenetic tree based on ML analysis with 26 Ulva chloroplast genomes Numbers at the nodes indicate bootstrap values. GenBank accession numbers for all reference sequences are provided. Chloroplast genome in bold was from sample U161. |
As shown in Fig. 4, the relative synonymous codon usage (RSCU) values were calculated and summarized with chloroplast genomes of U. linza and two ecotypes of U. prolifera respectively. In general, it was clearly indicated that the codon selection strategies in the three chloroplast genomes were extremely similar. Except for methionine and tryptophan (RSCU=1), most amino acids were exhibited to have codon bias. A total of 26 high frequency codons (RSCU > 1), including a stop codon, were identified with A/T ending as usual in Ulva (Cai et al., 2017), while the codons with negative bias (RSCU < 1) were prone to end with G/C. The results showed that the codon usage of the three genomes are extremely conservative without potential to provide resources for interspecific discrimination. Furthermore, we analyzed the genetic variations within the non-coding regions, including introns and gene spacer regions.

|
| Fig.4 RSCU of all 64 codons for protein-coding genes from three chloroplast genomes The three groups of data for each amino acid were U. prolifera (MZ571508, attached), U. prolifera (NC036137, floating), and U. linza (NC030312) from left to right. Ter: termination codon. |
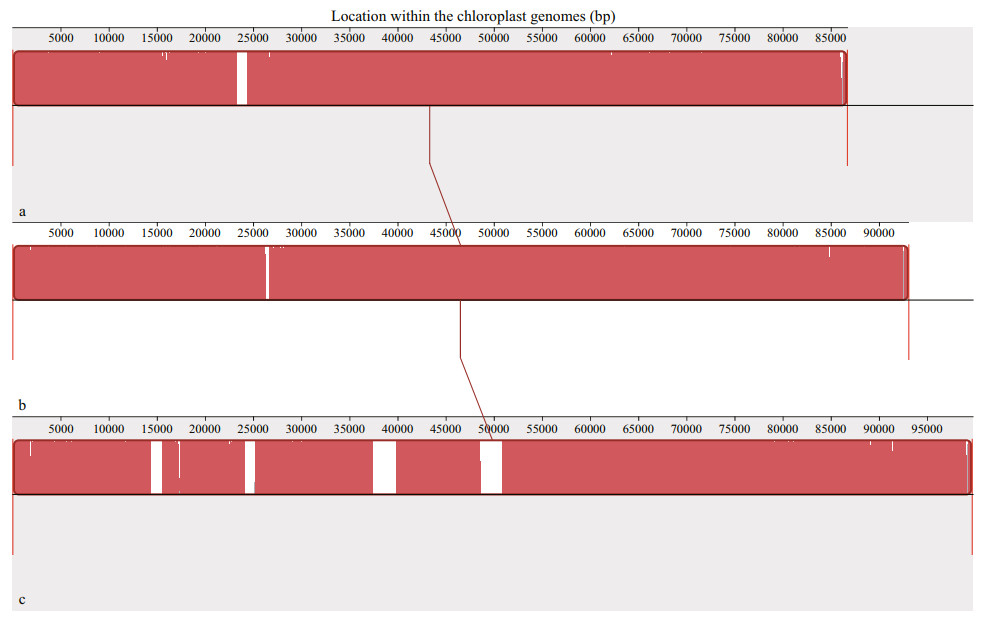
The collinearity analysis was conducted with these three chloroplast genomes. It was obviously shown that none of structural rearrangements such as inversions or translocations were detected among three genomes, and the orders of similarity sequences in the chloroplast genomes of U. linza and two ecotypes of U. prolifera were almost identical except for some slight variations such as insertions and deletions mainly located in the regions of introns or gene spacers (Fig. 5). Therefore, results of both the codon bias and collinearity analysis showed that U. linza had a very close genetic relationship with U. prolifera.
|
| Fig.5 Collinearity analysis among chloroplast genomes of U. linza and two ecotypes of U. prolifera a. U. linza (NC030312); b. U. prolifera (NC036137, floating); c. U. prolifera (MZ571508, attached). Local collinear blocks were shown as blocks with the same color. Blocks below the center line were aligned in reverse complementary orientation compared to the reference sequence and blocks above the center line were in forward orientation. |
To investigate the interspecific variations between U. linza and U. prolifera, these three chloroplast genomic sequences were compared using mVISTA. As shown in Fig. 6, plenty of variations were detected which were distributed in both the conserved non-coding sequences (CNS) and exon regions. By ignoring the intraspecific variations between the two ecotypes of U. prolifera, such as the psbB-psbC spacer region, only those signals which were identical between two ecotypes of U. prolifera but divergent between U. linza and U. prolifera, were further searched out to represent the interspecific variations between these two related species. A total of 454 SNPs, 131 indels and six structural variations were identified. In particular, three of the six structural variations were found to be longer than 1 000 bp. According to the position displayed on the X axis which was based on the chloroplast genome sequence of the attached ecotype of U. prolifera (MZ571508), these three regions of large structural variations were found to be located at psaB (3 kb–4 kb), petB (70.5 kb–71.8 kb), and psbB (91 kb–92 kb) respectively. Upon further analysis, each region was determined as an intron in the chloroplast genomes of U. prolifera, while it was a complete deletion in that of U. linza.

|
| Fig.6 Comparison of the chloroplast genome sequences among U. linza and two ecotypes of U. prolifera by mVISTA a. U. prolifera (MZ571508, attached); b. U. prolifera (NC036137, floating); c. U. linza (NC030312). Gray arrows on the top of the aligned sequences represent genes and their orientation. The pink regions are 'Conserved Non-Coding Sequences' (CNS) and the purple regions are exons. The percentages (50% and 100%) are the similarity among these sequences. |
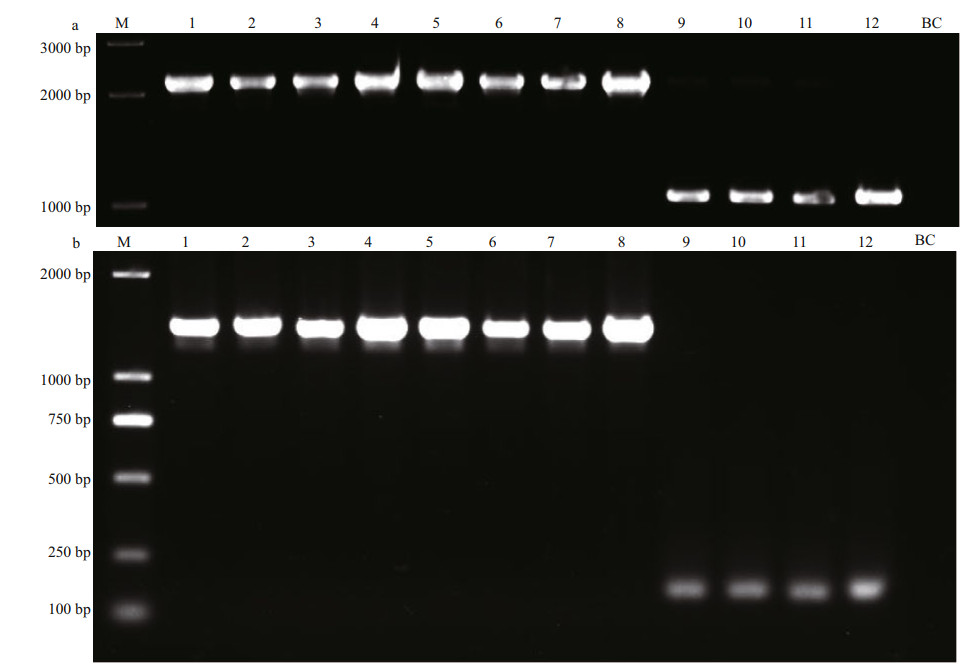
Dozens of pairs of primers were designed to target those interspecific variations between U. prolifera and U. linza which were located in either CNS or exon regions. After validation with PCR amplifications, those primers generating no products, polymorphic products, different products between two ecotypes of U. prolifera, or identical products between two related species, were all abandoned. Finally, two pairs of primers, which were designed to match the coding regions within psaB gene and petB gene respectively (Supplementary Table S2), were proved to be capable of generating species-specific signals to distinguish U. prolifera and U. linza (Fig. 7), and the sequences data have been uploaded to NCBI (Supplementary Table S3). The primers for psaB marker could amplify approximately 2 100-bp bands of the same size from both ecotypes of U. prolifera, while only about 1 000-bp bands can be amplified from U. linza. Similarly, the primers for petB marker could amplify an about 1 800-bp band from each of U. prolifera samples, whereas about 150-bp bands in U. linza samples. The ML phylogenetic trees for petB and psaB markers showed that all samples were clearly resolved into two clades, i.e., U. prolifera and U. linza, without significant genetic divergency in each clade (Supplementary Figs.S2–S3).
|
| Fig.7 PCR detection of psaB and petB markers a. amplification of psaB marker; b. amplification of petB marker. 1-4: U. prolifera (attached); 5-8: U. prolifera (floating); 9-12: U. linza. 1: QD240-1; 2: N155-17; 3: U246-22; 4: U161; 5: S096; 6: QD194-3; 7: N235-8; 8: N253-5; 9: 20-02-003; 10: U422-4; 11: U312-2; 12: QD233. M: Trans2K Plus II DNA Marker. BC: blank control. |
The phenotypic differentiations between the two ecotypes of U. prolifera have long been concerned, in terms of the morphology (Wang et al., 2010; Hiraoka et al., 2011; Gao et al., 2016; Ma et al., 2020), habitats (Ding et al., 2009), and transcription level of some key metabolism-related genes (He et al., 2019). Their significant differences in performances of reproductive isolation with U. linza were also described (Hiraoka et al., 2011). In particular, the genetic variations have also been revealed, by using inter-simple sequence repeat (ISSR) markers which were located throughout whole genomes (Zhao et al., 2011). A SCAR marker specific to the floating ecotype has been developed to find that this unique ecotype almost never formed a colonization population in the intertidal zone (Zhao et al., 2018). These findings implied the genetic differentiation between the two ecotypes of U. prolifera, which was confirmed to some extent by the comparative chloroplast genomic analysis in this study. In contrast, the results of all four tested molecular markers, especially 5S spacer, showed that all U. linza samples from different geographic populations were almost genetically identical, suggesting that the intraspecific genetic differences in U. linza were not significant. Therefore, in order to develop species-specific molecular markers for U. linza and U. prolifera, the influence of intraspecific differences, especially for U. prolifera that consisting of different ecotypes, should be fully considered.
In this study, the chloroplast genome from one representative attached population of U. prolifera was completely sequenced, and comparative analysis was performed with other chloroplast genomes from U. linza and the floating ecotype of U. prolifera. A strategy was proposed that only those signals of variation which were identical between two ecotypes of U. prolifera but divergent between U. linza and U. prolifera, were selected to develop the interspecific markers for U. linza and U. prolifera. Two candidate markers, i.e. psaB and petB, were validated to be capable of distinguishing these two related species. These new markers are expected to be used in surveys for Ulva species composition and green tide monitoring especially in the southern Yellow Sea. This sea area has experienced severe green tides for more than a decade (Yu et al., 2018). It was proposed that, the fouling green seaweeds on the nori rafts at Subei, in which both U. linza and U. prolifera were major members (Fan et al., 2015; Huo et al., 2015), provided the origin of biomass for the green tides in the Yellow Sea (Liu et al., 2009), and only the floating ecotype of U. prolifera finally succeed to be extremely dominant (Zhao et al., 2015). In addtion, it was suggested that the Ulva species composition and biomass in samples, including those fouling green seaweeds and the Ulva micro-propagules distributed in seawaters or surface sediments in this area, might contribute greatly to the interannual characteristics of the green tides (Song et al., 2015). Therefore, the new interspecific markers developed in this study, in combination with the existing floating ecotypes-specific marker, are expected to be able to characterise the detailed dynamic characteristics of the Yellow Sea green tide, and provided important data for effective risk mornitoring and management.
Organelle genomes contain abundant genetic resources, the mitochondrial genome sizes in Ulva vary between 55 kb to 88 kb, and the chloroplast genome sizes are 86 kb–119 kb. At present, genome sequences including 33 mitochondria and 26 chloroplasts from Ulva have been available in the GenBank database (https://www.ncbi.nlm.nih.gov), which were conducive to the development of molecular markers and used for inter- or intra-specific phylogenetic analysis. Recent studies showed that, for some widely distributed Ulva species, such as U. pertusa (synonym of U. australis) and U. compressa, a certain degree of intraspecific variations in organelle genomes have been detected among different geographic populations (Liu et al., 2017, 2020a; Cai et al., 2021). Since U. linza and U. prolifera also occurred worldwide, the organelle genome resources we provided in this study could contribute to validation or development of interspecific markers in future.
Moreover, in addition to the interspecific markers for U. linza and U. prolifera, the candidates of intraspecific markers specific to the floating ecotype of U. prolifera were also noted in this study. Novel organelle genome-derived markers could be developed from the chloroplast genomes and could be used together with the nuclear genome-derived SCAR marker for the ecological investigation of the green tide of Yellow Sea (Zhao et al., 2015).
5 CONCLUSIONIn this study, a chloroplast genome from one attached population of U. prolifera was completely sequenced, and comparative genome analysis was performed with other existing chloroplast genomes from U. linza and the floating ecotype of U. prolifera. The results showed that in spite of the high level of collinearity among three genomes, there were plenty of interspecific and intraspecific genetic variations. Two developed markers, psaB and petB, were shown to be able to distinguish these two closely related species and were applicable to more attached populations of U. prolifera from a wide range of geographical sources.
6 DATA AVAILABILITY STATEMENTThe genome sequence data of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession No. MZ571508. The datasets analyzed during the current study were available from the corresponding author on reasonable request.
Electronic supplementary material
Supplementary material (Supplementary Tables S1–S3 and Figs.S1–S3) is available in the online version of this article at https://doi.org/10.1007/s00343-022-2045-x.
Blomster J, Bäck S, Fewer D P, et al. 2002. Novel morphology in Enteromorpha (Ulvophyceae) forming green tides. American Journal of Botany, 89(11): 1756-1763.
DOI:10.3732/ajb.89.11.1756 |
Blomster J, Maggs C A, Stanhope M J. 1998. Molecular and morphological analysis of Enteromorpha intestinalis and E. compressa (Chlorophyta) in the British Isles. Journal of Phycology, 34(2): 319-340.
DOI:10.1046/j.1529-8817.1998.340319.X |
Blomster J, Maggs C A, Stanhope M J. 1999. Extensive intraspecific morphological variation in Enteromorpha muscoides (Chlorophyta) revealed by molecular analysis. Journal of Phycology, 35(3): 575-586.
DOI:10.1046/j.l529-8817.1999.3530575.x |
Cai C E, Gu K, Zhao H, et al. 2021. Screening and verification of extranuclear genetic markers in green tide algae from the Yellow Sea. PLoS One, 16(6): e0250968.
DOI:10.1371/journal.pone.0250968 |
Cai C E, Wang L K, Zhou L J, et al. 2017. Complete chloroplast genome of green tide algae Ulva flexuosa (Ulvophyceae, Chlorophyta) with comparative analysis. PLoS One, 12(9): e0184196.
DOI:10.1371/journal.pone.0184196 |
Chan P P, Lowe T M. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. In: Kollmar M ed. Gene Prediction. Humana, New York. p. 1-14, https://doi.org/10.1007/978-1-4939-9173-0_1.
|
Cui J J, Monotilla A P, Zhu W R, et al. 2018. Taxonomic reassessment of Ulva prolifera (Ulvophyceae, Chlorophyta) based on specimens from the type locality and Yellow Sea green tides. Phycologia, 57(6): 692-704.
DOI:10.2216/17-139.1 |
Darling A E, Mau B, Perna N T. 2010. ProgressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One, 5(6): el1147.
DOI:10.1371/journal.pone.0011147 |
Ding L P, Fei X G, Lu Q Q, et al. 2009. The possibility analysis of habitats, origin and reappearance of bloom green alga (Enteromorpha prolifera) on inshore of western Yellow Sea. Chinese Journal of Oceanology and Limnology, 27(3): 421-424.
DOI:10.1007/s00343-009-9277-x |
Duan W J, Guo L X, Sun D, et al. 2012. Morphological and molecular characterization of free-floating and attached green macroalgae Ulva spp. in the Yellow Sea of China. Journal of Applied Phycology, 24(1): 97-108.
DOI:10.1007/s10811-011-9654-7 |
Fan S L, Fu M Z, Wang Z L, et al. 2015. Temporal variation of green macroalgal assemblage on Porphyra aquaculture rafts in the Subei Shoal, China. Estuarine, Coastal and Shelf Science, 163: 23-28.
DOI:10.1016/j.ecss.2015.03.016 |
Frazer K A, Pachter L, Poliakov A, et al. 2004. VISTA: computational tools for comparative genomics. Nucleic Acids Research, 32(S2): W273-W279.
DOI:10.1093/nar/gkh458 |
Gao G, Zhong Z H, Zhou X H, et al. 2016. Changes in morphological plasticity of Ulva prolifera under different environmental conditions: a laboratory experiment. Harmful Algae, 59: 51-58.
DOI:10.1016/j.hal.2016.09.004 |
Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Research, 47(W1): W59-W64.
DOI:10.1093/nar/gkz238 |
Guidone M, Thornber C, Wysor B, et al. 2013. Molecular and morphological diversity of Narragansett Bay (RI, USA) Ulva (Ulvales, Chlorophyta) populations. Journal of Phycology, 49(5): 979-995.
DOI:10.1111/jpy.12108 |
Guiry M D, Guiry G M. 2021. AlgaeBase. World-wide electronic publication. Galway: National University of Ireland. https://www.algaebase.org/search/genus/detail/?genus_id=33.
|
Han W, Chen L P, Zhang J H, et al. 2013. Seasonal variation of dominant free-floating and attached Ulva species in Rudong coastal area, China. Harmful Algae, 28: 46-54.
DOI:10.1016/j.hal.2013.05.018 |
Hay den H S, Blomster J, Maggs C A, et al. 2003. Linnaeus was right all along: Ulva and Enteromorpha are not distinct genera. European Journal of Phycology, 38(3): 277-294.
DOI:10.1080/1364253031000136321 |
He Y, Ao Y, Yin Y, et al. 2019. Comparative transcriptome analysis between floating and attached Ulva prolifera in studying green tides in the Yellow Sea. Algal Research, 44: 101712.
DOI:10.1016/j.algal.2019.101712 |
Hiraoka M, Ichihara K, Zhu W R, et al. 2011. Culture and hybridization experiments on an Ulva clade including the Qingdao strain blooming in the Yellow Sea. PLoS One, 6(5): el9371.
DOI:10.1371/journal.pone.0019371 |
Hofmann L C, Nettleton J C, Neefus C D, et al. 2010. Cryptic diversity of Ulva (Ulvales, Chlorophyta) in the Great Bay Estuarine System (Atlantic USA): introduced and indigenous distromatic species. European Journal of Phycology, 45(3): 230-239.
DOI:10.1080/09670261003746201 |
Huo Y Z, Han H B, Shi H H, et al. 2015. Changes to the biomass and species composition of Ulva sp. on Porphyra aquaculture rafts, along the coastal radial sandbank of the Southern Yellow Sea. Marine Pollution Bulletin, 93(1-2): 210-216.
DOI:10.1016/j.marpolbul.2015.01.014 |
Kang J H, Jang J E, Kim J H, et al. 2019. Species composition, diversity, and distribution of the genus Ulva along the coast of Jeju Island, Korea based on molecular phylogenetic analysis. PLoS One, 14(7): e0219958.
DOI:10.1371/journal.pone.0219958 |
Kessler R W, Weiss A, Kuegler S, et al. 2018. Macroalgal-bacterial interactions: role of dimethylsulfoniopropionate in microbial gardening by Ulva (Chlorophyta). Molecular Ecology, 27(8): 1808-1819.
DOI:10.1111/mec.14472 |
Kuraku S, Zmasek C M, Nishimura O, et al. 2013. aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Research, 41(W1): W22-W28.
DOI:10.1093/nar/gkt389 |
Lagesen K, Hallin P, Rodland E A, et al. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Research, 35(9): 3100-3108.
DOI:10.1093/nar/gkm160 |
Leliaert F, Zhang X W, Ye N H, et al. 2009. Research note: identity of the Qingdao algal bloom. Phycological Research, 57(2): 147-151.
DOI:10.1111/j.1440-1835.2009.00532.x |
Leskinen E, Pamilo P. 1997. Evolution of the ITS sequences of ribosomal DNA in Enteromorpha (Chlorophyceae). Hereditas, 126(1): 17-23.
DOI:10.1111/j.l601-5223.1997.00017.x |
Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25(11): 1451-1452.
DOI:10.1093/bioinformatics/btp187 |
Liu D Y, Keesing J K, Xing Q G, et al. 2009. World's largest macroalgal bloom caused by expansion of seaweed aquaculture in China. Marine Pollution Bulletin, 58(6): 888-895.
DOI:10.1016/j.marpolbul.2009.01.013 |
Liu F, Melton III J T, Bi Y P. 2017. Mitochondrial genomes of the green macroalga Ulva pertusa (Ulvophyceae, Chlorophyta): novel insights into the evolution of mitogenomes in the Ulvophyceae. Journal of Phycology, 53(5): 1010-1019.
DOI:10.1111/jpy.12561 |
Liu F, Melton III J T, Lopez-Bautista J M, et al. 2020a. Multiple intraspecific variations of mitochondrial genomes in the green-tide forming alga, Ulva compressa Linnaeus (Ulvophyceae, Chlorophyta). Frontiers in Marine Science, 7: 714.
DOI:10.3389/fmars.2020.00714 |
Liu J L, Zhao X H, Kang X Y, et al. 2020b. Good news: we can identify Ulva species erupted in the Yellow Sea more easily and cheaply now. Conservation Genetics Resources, 12(3): 447-449.
DOI:10.1007/s12686-019-01114-x |
Luo R B, Liu B H, Xie Y L, et al. 2015. Erratum: SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience, 4: 30.
DOI:10.1186/s13742-015-0069-2 |
Ma Y Y, Zhao J, Xie W F, et al. 2020. Branching phenotype and plasticity in floating ecotype of Ulva prolifera (Ulvophyceae, Chlorophyta). Marine Sciences, 44(8): 98-105.
(in Chinese with English abstract) DOI:10.11759/hykx20200121001 |
Mantri V A, Kazi M A, Balar N B, et al. 2020. Concise review of green algal genus Ulva Linnaeus. Journal of Applied Phycology, 32(5): 2725-2741.
DOI:10.1007/s10811-020-02148-7 |
Meade J C, Shah P H, Lushbaugh W B. 1997. Trichomonas vaginalis: analysis of codon usage. Experimental Parasitology, 87(1): 73-74.
DOI:10.1006/expr.1997.4185 |
Melton III J T, Lopez-Bautista J M. 2021. Diversity of the green macroalgal genus Ulva (Ulvophyceae, Chlorophyta) from the East and Gulf Coast of the United States based on molecular data. Journal of Phycology, 57(2): 551-568.
DOI:10.1111/jpy.13120 |
Ogawa T, Ohki K, Kamiya M. 2013. Differences of spatial distribution and seasonal succession among Ulva species (Ulvophyceae) across salinity gradients. Phycologia, 52(6): 637-651.
DOI:10.2216/13-199.1 |
Qu X J, Moore M J, Li D Z, et al. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods, 15: 50.
DOI:10.1186/sl3007-019-0435-7 |
Shimada S, Yokoyama N, Arai S, et al. 2008. Phylogeography of the genus Ulva (Ulvophyceae, Chlorophyta), with special reference to the Japanese freshwater and brackish taxa. Journal of Applied Phycology, 20(5): 979-989.
DOI:10.1007/s10811-007-9296-y |
Song W, Han H B, Wang Z L, et al. 2019. Molecular identification of the macroalgae that cause green tides in the Bohai Sea, China. Aquatic Botany, 156: 38-46.
DOI:10.1016/j.aquabot.2019.04.004 |
Song W, Li Y, Fang S, et al. 2015. Temporal and spatial distributions of green algae micro-propagules in the coastal waters of the Subei Shoal, China. Estuarine, Coastal and Shelf Science, 163: 29-35.
DOI:10.1016/j.ecss.2014.08.006 |
Steinhagen S, Karez R, Weinberger F. 2019. Cryptic, alien and lost species: molecular diversity of Ulva sensu lato along the German coasts of the North and Baltic Seas. European Journal of Phycology, 54(3): 466-483.
DOI:10.1080/09670262.2019.1597925 |
Tamura K, Stecher G, Peterson D, et al. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30(12): 2725-2729.
DOI:10.1093/molbev/mst197 |
Tan I H, Blomster J, Hansen G, et al. 1999. Molecular phylogenetic evidence for a reversible morphogenetic switch controlling the gross morphology of two common genera of green seaweeds, Ulva and Enteromorpha. Molecular Biology and Evolution, 16(8): 1011-1018.
DOI:10.1093/oxfordjournals.molbev.a026190 |
Wang J F, Jiang P, Cui Y L, et al. 2010. Molecular analysis of green-tide-forming macroalgae in the Yellow Sea. Aquatic Botany, 93(1): 25-31.
DOI:10.1016/j.aquabot.2010.03.001 |
Xie W F, Mei X Y, Jiang P. 2019. Evaluation of tuf A for distinguishing dominant species (Ulva prolifera) of green tide in Yellow Sea. Environmental Science and Management, 44(12): 182-184.
(in Chinese with English abstract) |
Xie W F, Wu C H, Zhao J, et al. 2020. New records of Ulva spp.(Ulvophyceae, Chlorophyta) in China, with special reference to an unusual morphology of U. meridionalis forming green tides. European Journal of Phycology, 55(4): 412-425.
DOI:10.1080/09670262.2020.1740946 |
Yang J B, Tang M, Li H T, et al. 2013. Complete chloroplast genome of the genus Cymbidium: lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evolutionary Biology, 13: 84.
DOI:10.1186/1471-2148-13-84 |
Yu R C, Sun S, Yan T, et al. 2018. Progresses and perspectives on green-tide studies in the Yellow Sea. Oceanologia et Limnologia Sinica, 49(5): 942-949.
(in Chinese with English abstract) DOI:10.11693/hyhz20180700158 |
Zhang D, Gao F L, Jakovlić I, et al. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Molecular Ecology Resources, 20(1): 348-355.
DOI:10.1111/1755-0998.13096 |
Zhang J H, Liu C C, Yang L L, et al. 2015. The source of the Ulva blooms in the East China Sea by the combination of morphological, molecular and numerical analysis. Estuarine, Coastal and Shelf Science, 164: 418-424.
DOI:10.1016/j.ecss.2015.08.007 |
Zhang X W, Xu D, Mao Y Z, et al. 2011. Settlement of vegetative fragments of Ulva prolifera confirmed as an important seed source for succession of a large-scale green tide bloom. Limnology and Oceanography, 56(1): 233-242.
DOI:10.4319/lo.2011.56.L0233 |
Zhang Y, Wang Z F, Guo Y N, et al. 2021. Complete chloroplast genomes of Leptodermis scabrida complex: comparative genomic analyses and phylogenetic relationships. Gene, 791: 145715.
DOI:10.1016/j.gene.2021.145715 |
Zhao J, Jiang P, Liu Z Y, et al. 2011. Genetic variation of Ulva (Enteromorpha) prolifera (Ulvales, Chlorophyta)—the causative species of the green tides in the Yellow Sea, China. Journal of Applied Phycology, 23(2): 227-233.
DOI:10.1007/s10811-010-9563-1 |
Zhao J, Jiang P, Liu Z Y, et al. 2013. The Yellow Sea green tides were dominated by one species, Ulva (Enteromorpha) prolifera, from 2007 to 2011. Chinese Science Bulletin, 58(19): 2298-2302.
DOI:10.1007/s11434-012-5441-3 |
Zhao J, Jiang P, Qin S, et al. 2015. Genetic analyses of floating Ulva prolifera in the Yellow Sea suggest a unique ecotype. Estuarine, Coastal and Shelf Science, 163: 96-102.
DOI:10.1016/j.ecss.2015.05.027 |
Zhao J, Jiang P, Qiu R, et al. 2018. The Yellow Sea green tide: a risk of macroalgae invasion. Harmful Algae, 77: 11-17.
DOI:10.1016/j.hal.2018.05.007 |
 2022, Vol. 40
2022, Vol. 40


