Institute of Oceanology, Chinese Academy of Sciences
Article Information
- HAO Wenjin, WANG Lei, LI Fan, SUN Tingting, PENG Saijun, LI Yongxue, ZHAO Jianmin, DONG Zhijun
- Bacterial communities associated with hydromedusa Gonionemus vertens in different regions in Chinese coastal waters
- Journal of Oceanology and Limnology, 40(4): 1530-1543
- http://dx.doi.org/10.1007/s00343-021-1036-7
Article History
- Received Feb. 7, 2021
- accepted in principle Jun. 9, 2021
- accepted for publication Aug. 25, 2021
2 Muping Coastal Environment Research Station, Yantai Institute of Coastal Zone Research, Chinese Academy of Sciences, Yantai 264003, China;
3 Shandong Key Laboratory of Marine Ecological Restoration, Shandong Marine Resource and Environment Research Institute, Yantai 264006, China;
4 Center for Ocean Mega-Science, Chinese Academy of Sciences, Qingdao 266071, China
Jellyfish blooms are a growing concern from marine aquaculture industry, as they have been associated with an increasingly number of mortality events in Atlantic salmon farming, which has resulted in economic losses and fish welfare issues (Småge et al., 2018). In recent years, sudden fish mortalities in marine aquaculture were reported from Asia, Australia, and North and South America as results of episodic blooms of different jellyfish species (Bosch-Belmar et al., 2017).
Gonionemus vertens (Hydrozoa, Limnomedusae) is a highly toxic cryptogenic clinging jellyfish which has outbreaks in coastal areas around the world and is considered an invasive species (Rodriguez et al., 2014; Gaynor et al., 2016; Govindarajan and Carman, 2016; Govindarajan et al., 2017; Marchessaux et al., 2017). G. vertens was introduced to the Mediterranean Sea, and has now invaded into the northern Europe (Bakker, 1980), the western North Atlantic (Govindarajan and Carman, 2016), and the western South Atlantic (Rodriguez et al., 2014), indicating that it can readily travelled and invade into new areas (Schuchert, 2016). Invasive species can have harmful impacts on local native taxa and disrupt normal ecosystem functioning (Gallardo et al., 2016; Govindarajan et al., 2017). In addition, they can also have direct negative impacts on human health (Ruiz et al., 2000; Pyšek and Richardson, 2010). The hydromedusae have a potent sting for causing severe pain and other symptoms in humans (Govindarajan and Carman, 2016; Marchessaux et al., 2017), and can be lethal to their predators (Carman et al., 2017).
A great number of jellyfish can clog fish cages by tidal movement, which would obstruct water exchange, resulting in oxygen depletion and jellyfish toxin buildup. Moreover, the stuck jellyfish could be torn off by ropes or wires of fish cages into toxic nematocyst-containing pieces that can pass through the mesh of the fish cages (Delannoy et al., 2011). Nematocysts could still be released from the pieces, which may be taken into or adhere to the fish, leading to severe lesions or fatal, especially when gills are infected (Ferguson et al., 2010). Jellyfish toxins are the most potent of all venoms, as they have cytotoxic, neurotoxic, cardiotoxic, hemolytic, dermatonecrotic, immunogenic, and inflammatory effects (Mariottini et al., 2008). If fish did not die directly from the toxins, they would succumb within a few hours due to respiratory failure or later from secondary bacterial infections to the body and the gills, caused by opportunistic bacteria such as Flavobacterim and Vibrios (Ferguson et al., 2010).
Significant species-specific bacterial differences between native jellyfish (Aurelia aurita) and invasive gelatinous zooplankton (Blackfordia virginica and the comb jelly Mnemiopsis leidyi) were confirmed, and the latter mainly consisted of Mycoplasma and Vibrio (Jaspers et al., 2020). The microbiome associated with jellyfish Rhizostoma pulmo is identified in three major prokaryotic genera (Spiroplasma, Mycoplasma, and Wolinella), which are considered bacterial vectors and potential hazards for marine animals and human health (Basso et al., 2019; Clinton et al., 2020). Cnidarian jellyfish may also be key carriers for bacterial pathogens as Tenacibaculum maritimum that was isolated from Pelagia noctiluca was implicated in cultured marine fish gill disease (Delannoy et al., 2011; Fringuelli et al., 2012). T. maritimum has been reported to infect gills damaged by jellyfish venom (Delannoy et al., 2011). The pathogenesis and natural reservoir of this pathogen has not yet been clarified, and the associated lesions are characterized by necrosis in the mouth, head, fins, and gills (Delannoy et al., 2011). Pathogenesis of the lesions is thought to be the result of the proteolytic activity of extracellular toxins (Delannoy et al., 2011). The disease, which is recognized worldwide, is considered an important threat to aquaculture (Toranzo et al., 2005). However, the medusae were collected far away from sea-cages of infected salmon; since these jellyfish are small enough to pass the cages, it was impossible to determine the original source of the bacteria (Delannoy et al., 2011).
In the present study, we investigated the bacterial communities associated with G. vertens from two different environments to determine if this hydromedusa species is a direct potential host of marine agriculture pathogen. We aimed to analyze the bacterial community compositions associated with hydromedusa G. vertens in both marine agriculture and natural environments using 16S rRNA sequencing, attempt to determine the diversity, composition, and potential functions of the bacterial communities, and to compare the associated bacterial communities from different geographic regions. This study provides information regarding the associated bacterial communities of hydromedusae G. vertens outbreaks for the maintenance of environment and marine organisms.
2 MATERIAL AND METHOD 2.1 Animals collection and preparationGonionemus vertens was sampled in two different environments (Fig. 1): sea cucumber culture areas in Dalian, Liaoning (coded GVDL; 39°34′21″N, 122°22′47″E) and Dongying, Shandong (coded GVDY; 37°95′21″N, 119°12′05″E), and a natural marine environment in the Sishili Bay, Yantai, Shandong (coded GVYT; 37°56′09″N, 121°52′02″E), using a 500-μm dip net towed onboard a research vessel with long-handled bucket. In each station, 10 intact G. vertens specimens were taken and immediately placed into sterile seawater (pre-filtered by 0.2-μm pore size filter and autoclaved) baths for 3 h to clear their guts and remove loosely associated microorganisms. Each specimen was rinsed three times with sterile seawater before further analysis, stored at -20 ℃, and freeze-dried before DNA extraction. Seawater samples (500 mL) collected from each location at the same time as seawater sample (coded as SWDL, SWDY, and SWYT, respectively to the three areas of G. vertens sampling) were pre-filtered by 3-μm pore size filters (TCTP, 47 mm, Millipore, Germany) and then sequentially filtrated through 0.2-μm pore size filter (Millipore Corporation, Bedford, MA) with three parallels to obtain free-living bacterial community from water column for comparison to the jellyfish-associated bacteria (Daniels and Breitbart, 2012). The bacterial biomass from the filters was stored at -80 ℃ until further processing.
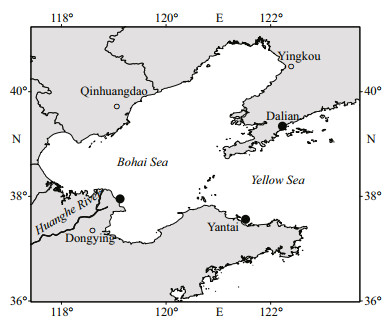
|
| Fig.1 Sampling locations in the Chinese coastal waters |
Total genomic DNA from the bacterial community associated with each individual G. vertens were extracted from freeze-dried tissues using cetyltrimethyl-ammonium bromide (CTAB) as per Hao et al. (2015) with slight modifications. The filter membranes of each seawater sample were applied to extract the total DNA from the seawater samples using a PowerWater DNA Isolation Kit (MOBIO, USA) according to the manufacturer's instructions. The hypervariable regions V4–V5 of the bacteria 16S rRNA gene were amplified with specific primers: 515F (5′-GTG CCA GCM GCC GCG GTA A-3′) and 907R (5′-CCG TCA ATT CCT TTG AGT TT-3′). All PCR reactions were carried out using the Phusion High-Fidelity PCR Master Mix (New England Biolabs) with 0.2 μmol/L of each primer and 10-ng template DNA. Thermal cycling consisted of initial denaturation at 98 ℃ for 1 min, followed by 30 cycles of denaturation at 98 ℃ for 10 s, annealing at 50 ℃ for 30 s, and elongation at 72 ℃ for 30 s, followed by 72 ℃ for 5 min (Gong et al., 2019). All PCRs were performed in triplicate, and no-template controls were included in all steps of the process. The PCR products were detected by electrophoresis in a 2% (w/v) agarose gel. The PCR amplicons of each sample that produced bright bands were mixed in equal density ratios and purified with GeneJETTM Gel Extraction Kit (Thermo Scientific). The amplicon libraries were generated using Ion Plus Fragment Library Kit 48 rxns (Thermo Scientific) and sequenced using the IonS5TMXL platform at Novogene Bioinformatics Technology Co., Ltd. (Beijing, China).
2.3 Bioinformatics and Statistical AnalysesThe analysis was conducted by QIIME2docs along with customized program scripts (https://docs.qiime2. org/2019.1/). Briefly, raw FASTQ files were demultiplexed using the QIIME2 demux plugin based on the unique barcodes. Demultiplexed sequences from each sample were quality filtered and trimmed, de-noised, merged, and then the chimeric sequences were identified and removed using the QIIME2 dada2 plugin to obtain the feature table of the amplicon sequence variant (ASV) (Gong et al., 2019). The QIIME2 feature-classifier plugin was then used to align the ASV sequences to a pre-trained SILVA database (trimmed to the V4–V5 region bound by the 515F/907R primer pair) to generate the taxonomy table (Bokulich et al., 2018). Any contaminating mitochondrial and chloroplast sequences were filtered using the QIIME2 feature-table plugin. Linear discriminant analysis (LDA) effect sizes (LEfSe), accounts for both bioconsistency and significance, were used to identify the significant differences in the bacterial taxa between the sea cucumber breeding area (GVDL and GVDY) and the natural marine environment (GVYT). Diversity metrics were calculated using the core-diversity plugin within QIIME2. Feature level alpha diversity indices, such as observed operational taxonomic units (OTUs), Chao1 richness estimator, Shannon diversity index, and faith_pd were calculated to estimate the microbial diversity within an individual sample (Gong et al., 2019). Beta diversity distance, including Bray Curtis and Weighted UniFrac were measured to investigate the variation of the bacterial communities structure among all samples, and visualized using principal coordinate analysis (PCoA). The Venn diagram was applied to screen out common OTU among different sample groups. Major bacterial communities were filtered with the relative abundant higher than 1% from these common OTU. Functional diversity of the bacterial communities associated with jellyfish G. vertens was predicted using PICRUSt (Langille et al. 2013). OTUs were assigned with QIIME's command "pick_closed_otus" with 97% similarity in Greengenes13.5 database. Then, the predicted functions were blasted to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database at group levels 1 and 2, and statistical differences among groups were analyzed using test package of R language. ANOVA+Duncan and Dunn tests were conducted to analyze whether there was significant difference in the prediction function of microbial community among groups, unless specified above, the default parameters were used in the analysis.
3 RESULT 3.1 Sequencing and classificationPCR products for the V4–V5 regions were sequenced using the single-end method with the IonS5TMXL platform, and this generated a total of 3 060 720 raw tags representing 39 samples, with individual reads ranging from 53 229 to 93 790. After qualification and removal of the chimeras from the raw tags, a total of 2 920 866 validated sequence reads, and 74 894 for each sample on average (ranging 51 493–89 956), and an average of 370 bp, were obtained. The effective tags of all samples were grouped into 58 278 OTUs based on 99% identities. Through normalized processing, a total of 4 039 OTUs were retained for all further downstream analyses. The trend of the rarefaction curves suggested that there was sufficient sampling of the bacterial communities, as it showed that each sample was different (Supplementary Fig.S1 & Table S1). This indicated that the bacterial diversity was well revealed by the high sequence number per sample, which was enough to acquire the majority of the 16S rRNA gene sequences.
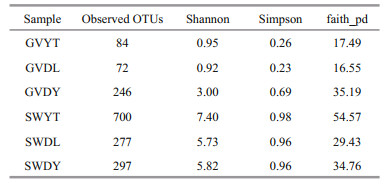
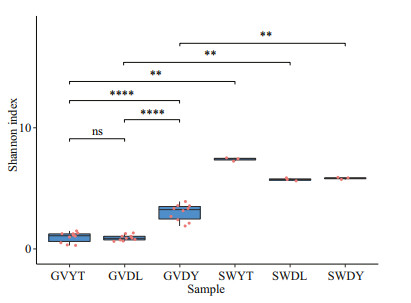
3.2 Analysis of alpha diversityThe alpha diversity of all samples is displayed in Table 1. The lowest OTUs were 72 in the GVDL samples and the highest were 246 in the GVDY samples among jellyfish samples. Moreover, the Shannon index of the GVDY samples was the highest (3.00). Compared with the natural environment samples, the SWYT sample had the highest value (7.40). We found that SWYT also had the highest Simpson indices (0.98) and faith-pd indices (54.57). According to the multiple comparisons based on Wilcox test, we found that the Shannon index of the GVDY was significantly different from that of GVDL and GVYT (P<0.001). In addition, the alpha diversity of the jellyfish samples at three locations (GVDL, GVDY, and GVYT) were significantly different from those of the seawater samples (SWDL, SWDY, and SWYT), respectively (Fig. 2).
|
|
| Fig.2 Alpha diversity of the bacterial communities associated with G. vertens and the environment seawater, represented by the Shannon index accompanied by multiple comparisons based on the Wilcox test **: P<0.01; ****: P<0.001; P-values represent the results from the Wilcox test. |
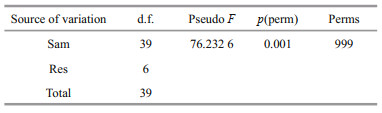
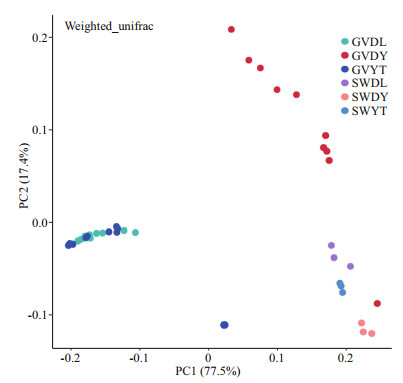
Principal coordinate analysis (PCoA) was performed to visualize and compare the structure of the bacterial communities among different samples. The PCoA results based on weighted unifrac distances demonstrated that the jellyfish samples (GVDL, GVDY, and GVYT) and seawater samples (SWDL, SWDY, and SWYT) tended to separate according to PC1 (77.5%) and PC2 (17.4%) (Fig. 3). The bacterial communities associated with GVDL clustered with those from GVYT, but they were all separated from the corresponding seawater samples. The PERMANOVA main test indicated significant differences among the bacterial communities associated with G. vertens among the three different regions (P=0.001; Table 2), where pair-wise comparisons indicated that the bacterial community associated with the GVDY was significantly different from the communities associated with GVDL and GVYT (Supplementary Table S2; P=0.001).
|
|
| Fig.3 Principal coordinate analysis (PCoA) presenting the bacterial communities associated with G. vertens and seawater from different locations based on weighted unifrac metrics |
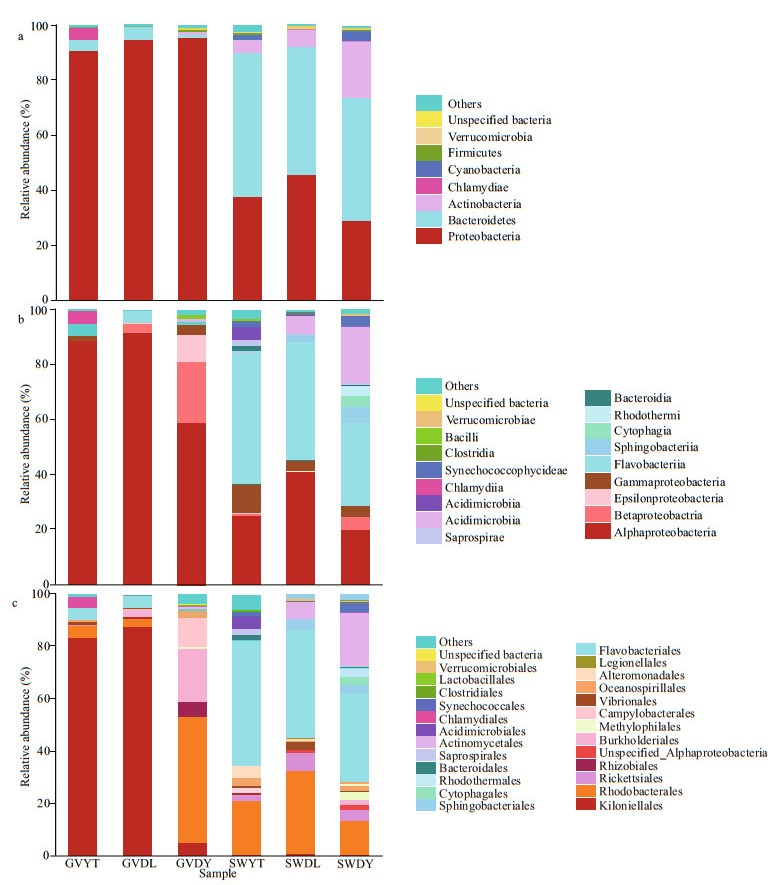
The relative abundances of the identified taxa from the phylum to order were reported as stacked bar plots for each analyzed sample (Fig. 4a–c). Only taxa with a relative abundance equal to or higher than 1% were plotted, and the rest was collapsed into "other".
|
| Fig.4 Stacked bar-plots of relative abundances of the bacterial communities compositions associated with jellyfish G. vertens and the environment seawater at three different locations at phylum (a), class (b), and order (c) levels Only the taxa with a relative abundance equal or higher than 1% were plotted, and the rest was grouped into "others". |
Proteobacteria dominated the observed sequences at the phylum level for jellyfish samples, representing 95.31%, 95.08%, and 90.95% of the total number of species in GVDL, GVDY, and GVYT, respectively, followed by Bacteroidetes accounting for 4.35%, 1.18%, and 4.01% of GVDL, GVDY, and GVYT, respectively. Furthermore, Bacteroidetes were found the predominant phylum in SWYT (52.25%), SWDL (46.06%), and SWDY (44.53%), followed by proteobacteria in the SWDL (45.89%), SWYT (37.51%), and SWDY (29.27%). Meanwhile, Actinobacteria levels were also high in the SWDY, SWDL, and SWYT samples, accounting for 20.62%, 6.34%, and 4.89%, respectively, followed by Cyanobacteria for 3.83% of the SWDY (Fig. 4a).
Within Proteobacteria, Alphaproteobacteria were the most dominant group (91.42%) at the class level in the GVDL sample, followed by GVYT (88.63%) and GVDY (59.33%). Flavobacteriia made up a small portion of the GVDL (4.3%), GVYT (3.9%), and GVDY (0.55%). In addition, the level of Betaproteobacteria was high in GVDY samples (22.17%), followed by Epsilonproteobacteria (9.95%) and Gammaproteobacteria (3.24%) (Fig. 4b). Concerning the natural environment seawater samples, Flavobacteriia occupied a large part of the relative abundance of SWYT (46.01%), SWDL (42.5%), and SWDY (31.55%) (Fig. 4b). Alphaproteobacteria levels were also high in SWDL (40.82%), SWYT (24.67%), and SWDY (19.82%), while the Actinobacteria made up 20.28% of SWDY (Fig. 4b).
Kiloniellales was the most abundant group in the GVDL (87.59%) and GVYT (83.98%) (Fig. 4c). In GVDY, Rhodobacterales and Burkholderiales were found the most dominant group accounting for 48.75% and 21.98%, respectively, followed by Campylobacterales (9.95%). In addition, Flavobacteriales and Rhodobacterales were dominant in the samples of seawater samples of SWDL (42.50% and 32.32%), SWDY (31.55% and 13.11%), and SWYT (46.01% and 20.81%). Actinomycetales accounted for 20.28% in SWDY, while Rickettsiales appeared in all SWDL, SWDY, and SWYT samples, accounting for 6.58%, 4.35%, and 1.78%, respectively (Fig. 4c).
3.5 LEfSe analysis revealed specific bacterial biomarkersBased on the above analyses of the diversity, similarity, and compositions of the bacterial communities associated with G. vertens among three locations, linear discriminant analysis (LDA) and the effect sizes (LEfSe) were applied to reveal the bacterial bioindicators that were significantly associated with G. vertens in the two different environments at the genus level. We identified 31 unique bacterial biomarkers (Fig. 5) in the bacterial communities associated with G. vertens among the three locations using the LEfSe analysis (LDA score >4 and P<0.05). Five bacterial taxa, including Rhodobacteraceae and Oxalobacteraceae, and its two members (i.e., Pseudoruegeria and Polynucleobacter) were detected as biomarkers in GVDL. Thirteen different taxa were identified as discriminant biomarkers of GVDY. Six and three out of the nine biomarkers identified in GVDY belong to Alphaproteobacteria (e.g., Octadecabacter and Labrenzia) and Epsilonproteobacteria (e.g., Arcobacter), respectively. In addition, Pseudonocardia belong to Actinobacteria, and the biomarkers in the GVYT included Streptococcaceae, and Polaribacter belonging to Bacteroidetes, and Vibrio and Roseobacter belonging to Proteobacteria.

|
| Fig.5 LDA effect size of the LEfSe analysis to identify potential bacterial bioindicators present at the genus level in G. vertens in different environments (LDA score >4 and P<0.05) |
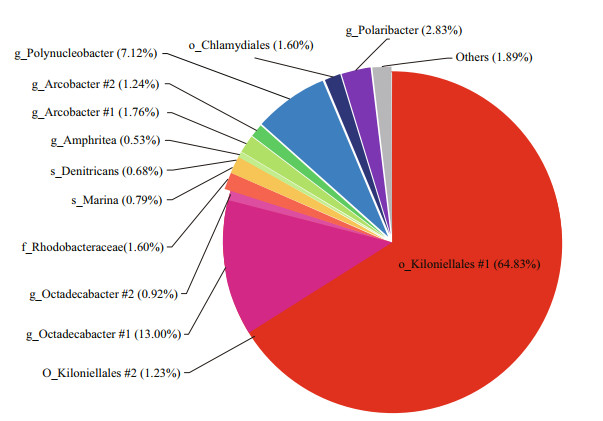
We further analyzed the core microbiome fractions from the three locations. The shared and unique OTUs among different samples were shown in the Venn graphs (Supplementary Fig.S2). There were 2 405 OTUs in total, which accounted for 59.54% of the total 16S rRNA gene reads of G. vertens. The OTU number drastically dropped to 55 when a restriction of each OTU (>1%) was observed in all locations of this study, and the 55 shared OTUs were regarded as the core OTUs. However, when only those with a relative abundance higher than the threshold value of 0.5% were considered, the number of core OTUs dropped rapidly to 14. According to our results, the major community was highly abundant with Kiloniellales (64.83% and 1.23%) and Octadecabacter (13.00% and 0.92%, respectively) (Fig. 6). The predominant colonizers were assigned to Polynucleobacter (7.12%), Polaribacter (2.83%), Rhodobacteraceae (1.60%), Arcobacter (1.76% and 1.24%), and Chlamydiales (1.6%) (Fig. 6).
|
| Fig.6 The major species bacterial communities (>0.5% relative abundance) of G. vertens among the three locations higher than 0.5% in relative abundance |
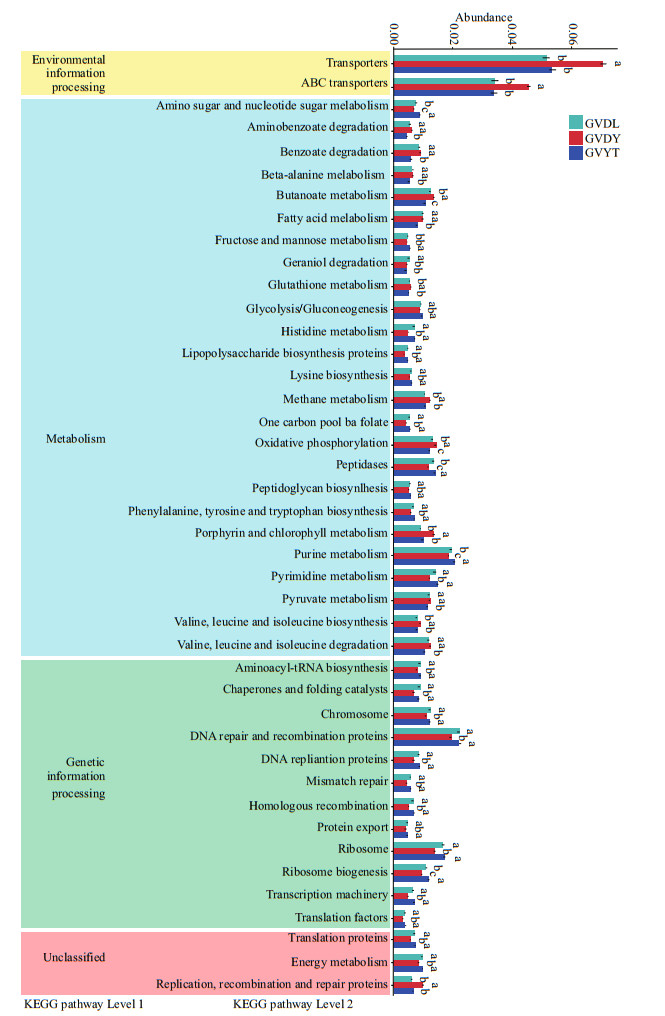
Using the Kyoto Encyclopedia of Genes and Genomes ortholog pathways (Langille et al., 2013), the KEGG functions of the identified bacteria were determined to be significantly (P<0.05) among three locations (Fig. 7). The results showed four potential functional categories including environmental information processing, metabolism, genetic information processing, and unclassified, respectively. There were 25 pathways identified that were involved in metabolism, and purine metabolism, peptidases, oxidative phosphorylation, butanoate metabolism, and amino sugar and nucleotide sugar metabolism were different among the three locations. In genetic information processing, DNA repair and recombination proteins and ribosome biogenesis were different between the GVDL, GVYT, and GVDY.
|
| Fig.7 The relative abundance of predicted KEGG functional pathway profiles based on PICRUSt among three locations The ordinate is the functional pathway at the second classification level of KEGG, and the left is the first level functional pathway to which the second path belongs; different colors represent different groups. If here is the same letter above two groups, the difference is not significant; otherwise, the difference is significant. |
Previous studies characterizing the bacteria associated with marine invertebrates focused largely on benthic organisms such as corals and sponges. They all contain microbiota that are distinct from the ambient water, and these bacteria play critical roles in host ecology (Cleary et al., 2016). In the present study, we characterized the bacterial communities associated with hydromedusa G. vertens that were collected from three different locations: two sea cucumber mariculture environments (Dongying and Dalian) and a natural marine environment (Sishili Bay, Yantai). The bacterial communities associated with G. vertens had a lower diversity as indicated in the 16S rRNA gene sequencing than that of the environmental seawater (Fig. 2 & Table 1). The bacterial communities associated with jellyfish from Dongying had the highest number of observed OTUs (246) and the highest Shannon index (3.00) when compared to the other two locations (Table 1). Cortés-Lara et al. (2015) and Viver et al. (2017) reported a reduced diversity of the microbiota associated with the gastric cavity of the jellyfish Cotylorhiza tuberculata. As clarified by Tinta et al. (2019) and Daley et al. (2016), the lower diversity in the microbial communities was associated with the invasive hydrozoan Nemopsis bachei and the cosmopolitan scyphozoan Aurelia aurita hosted taxon-specific bacterial groups. The bacterial community associated with ctenophore contained less bacterial OTUs and lower diversity communities than that in water column (Daniels and Breitbart, 2012). The low diversity of the jellyfish-associated bacterial communities differed from the trend previously observed in corals and sponges, in which bacterial diversity was typically higher than that of the surrounding water column (Rohwer et al., 2002; Taylor et al., 2007; Lee et al., 2011). Although these gelatinous animals are primarily composed of water, it has previously been reported that their associated bacterial communities are strikingly different from the surrounding water column (Daniels and Breitbart, 2012; Hao et al., 2015; Weiland-Bräuer et al., 2015; Daley et al., 2016; Lee et al., 2018; Tinta et al., 2019). In the present study, the bacterioplankton communities both from the natural environment (Sishili Bay) and the aquiculture area (the holothurian mariculture areas in Dongying and Dalian) had high diversity (observed OTUs, Shannon, and Simpson index) and were significantly distinct from the jellyfishassociations (P=0.001). The bacterial communities obtained from the natural environment (Sishili Bay) had higher observed OTUs (700) and Shannon's indexes than those in the sea cucumber mariculture areas in Dongying (297) and Dalian (277). The ANOSIM and PERMANOVA P-values indicate no overlap among the bacterial communities from jellyfish and from surrounding water.
The bacterial communities associated with G. vertens from the holothurian mariculture area (GVDL) and natural environment (GVYT), collected in the Yellow Sea, mainly consisted of Alphaproteobactereia. The communities from the holothurian mariculture area (GVDY) from the estuary area in the Bohai Sea were composed of Alphaproteobacteria, Betaproteobacteria, and Epsilonproteobacteria. The Huanghe River estuary and coastal habitats of the Bohai Sea are some of the most productive marine ecosystems on earth and suffer greatly from upstream and coastal anthropogenic pollution. Such area are extremely dynamic with steep physicochemical gradients due to the variability of the freshwater input and geomorphology. Due to the differences in geographic environments and ecological conditions among different sea areas, the microbial community structures and characteristics are diversified (Webster et al., 2015). Many abiotic factors shape the microbial distribution patterns, such as salinity (Webster et al., 2015), temperature (Lindh et al., 2013), pH (Wang et al., 2015a), depth (Walsh et al., 2016), nutrient status (Fodelianakis et al., 2014), spatial factors (Martiny et al., 2006), biological resources, and the marine environment (Wei et al., 2016). Differentiated gradients control the bacterial diversity, abundance, and biogeographic distribution, in which microbes are crucial for maintaining functional and structural balances in estuarine and open-sea ecosystems through active biogeochemical cycling and complex food webs (Webster et al., 2015; Wei et al., 2016). It was demonstrated in this study that spatial separation and environmental condition decided the bacterial communities and they were distinctly different in Dongying and Dalian in the Bohai Sea and the Yellow Sea, respectively, whereas in the jellyfish that were collected from the sea cucumber culture ponds in different sea areas, they had similar structures in Dalian and Yantai both in the Yellow Sea area. It has been reported that the intestinal flora of the sea cucumber Apostichopus japonicus is influenced by specific geographical environments of the Yellow Sea and Bohai Sea Ye et al., 2018). However, Hao et al. (2015) reported that a common member of the bacterial community of ctenophora that was closely related to Marinomonas species, occurred in different regions, e.g., Tampa Bay, the Gulf of Mexico, USA, the Gullmar fjords of Sweden, and German Bight in Germany. There has been much controversy concerning vertical and horizontal transmission of these associated microbial community (Martiny et al., 2006). Mixed strategies combine the best horizontal and vertical transmission modes and play an important role in the evolution and ecology of host-symbiont relationships (Russell et al., 2017; Bernasconi et al., 2019b). Environmental changes in physical disturbances, seasonality, and climate affect microbial symbionts composition directly or via the physiological responses of the host, which, in turn, lead to changes in the conditions of boundary layers microhabitat (Wahl et al., 2012). For instance, the offspring of host can directly benefit the symbionts through vertical transmission. Unfortunately, this may have adverse effects if the transmitted association is not optimal in different environment conditions from those of their parents. Meanwhile, horizontal transmission may help the host to take over diverse associations well fitted to the specific environment (Byler et al., 2013). It has been shown that there is a mechanism for microbial community isolation and differentiation in the marine environment. Ocean currents, geographical distance, seafloor landform, life history, differential genetic drift, and sharp changes in temperature and salinity could affect gene flow, resulting in isolation and genetic differentiation of different microbial species (Webster et al., 2010). Therefore, different jellyfish species who can use both strategies (horizontal transmission and vertical transmission) are more likely to have physiological and evolutionary advantages than those strictly relying on one or another (Bernasconi et al. 2019a).
Gammaproteobacteria, Bacteroidetes, and Alphaproteobacteria were found dominated in the bacterial sequences of hydromedusa Nemopsis bachei (Daley et al., 2016). Concerning the bacterial communities associated with G. vertens in Dongying, the most abundant alphaproteobacterial OTUs inhabiting jellyfish were assigned to orders Rhodobacterales (48.75%), Burkholderiales (21.98%), Campylobacterales (9.95%), Rhizobiales (5%), and Oceanospirllales (2%). In contrast, the bacterial communities associated with G. vertens from Yantai and Dalian were very similar, and dominated by Kiloniellales. Moreover, only 5% of Kiloniellales were present in jelly-associated community in Dongying station. Cleary et al. (2013) found that three OTUs assigned to Kiloniellales took more than 63% of the total bacterial community of sponge in marine lakes in the Kakaban and Maratua islands in Indonesia. Afterwards, they compared the bacterial communities in two jellyfish species (the 'golden' jellyfish Mastigias cf. papua, and the box jellyfish Tripedalia cf. cystophora) from marine lakes in the Berau region in the northeastern Borneo, Indonesia, and found that they were dominated by OTUs assigned to Gammaproteobacteria, Mollicutes, Spirochaetes, and Alphaproteobacteria. The most abundant alphaproteobacterial OTUs were assigned to orders Kiloniellales and Rhodobacterales. OTUs assigned to the order Kiloniellales had their greatest relative abundance in Tripidelia hosts (Cleary et al., 2016). Kiloniellales were rare or absent in the seawater samples from the natural environment in the present study. Kiloniella was first reported by Wiese et al. (2009) as a novel alphaproteobacterium, and was isolated from marine macroalga. The genus Kiloniella represents the type of the new family Kiloniellaceae fam. nov. and order Kiloniellales ord. nov. (Wiese et al., 2009). Phylogenetically, the genus Kiloniella is related to the type strains of three Thalassospira species (88.9%–89.2%), which have been described previously in cnidaria-associated bacterial studies (Cortés-Lara et al., 2015; Hao et al., 2015). Kiloniellales is mesophilic and chemoheterotrophic aerobe with the potential for denitrification, and it displays a typical marine growth response (Wiese et al., 2009). All members of the Kiloniellales were found in quite different marine habitats and were obtained from sponges (Jang et al., 2015), shrimp (Wang et al., 2015b), spider crab (Gerpe et al., 2017), and algae (Si et al., 2017).Wiese et al. (2009) provided genome insights into several metabolic properties, such as carbon and sulfur metabolism, and indicated the potential for denitrification and biosynthesis of the secondary metabolites of Kiloniella.
In the present study, only a few Vibrio and Chlamydiae were detected as biomarkers in Yantai station where G. vertens were collected from a natural marine environment. We did not detect any pathogen sequences in the bacterial communities associated with G. vertens in the sea cucumber culture pond in the Dongying and Dalian stations. Hydrozoan siphonophore Muggiaea atlantica and microscopic hydromedusae Phialella quadrata and Solmaris corona had crucial impact on fish mortality in Irish and Scottish fish farms (Baxter et al., 2012; Bosch-Belmar et al., 2017). In parallel, it has been hypothesized that ctenophores and jellyfish can serve as vectors for fish pathogens, and recent findings have revealed that bacteria associated with hydromedusa served as the source of secondary infections in farmed salmon (Ferguson et al., 2010; Delannoy et al., 2011). Schuett and Doepke (2010) reported that the tentacles of Hydrozoa Tubularia indivisia were associated with potential endobiotic pathogenic bacteria, such as Cobetia marina, Colwellia aestuarii, Endozoiciminas elysicola, Vibrio aestuarianus, Bacillus subtilis, and Ilyobacter psychrophilus (Schuett and Doepke, 2010). The most abundant sequences were affiliated with the genus Tenacibaculum, in which 20 species have been isolated from marine environments. Five species were from water and four from sediment samples, others species, such as T. crassostreae and T. adriaticum were associated with healthy bryozoans, sea anemones, oysters, sponges, and green algae (Ferguson et al., 2010; Viver et al., 2017). In particular, T. maritimum is a known fish pathogen that has been detected in Hydrozoa M. atlantica (Fringuelli et al., 2012) and P. quadrata (Ferguson et al., 2010), and is implicated in farmed fish gill disease. However, we did not detect any sequence similarity with this species in our study. The impact of G. vertens on mariculture, mainly its toxic stings that can cause neuroparalysis and anaphylactic shock, and the effects of the pathogenic bacteria (e.g. the cultivation and specific phylogentic analysis of known pathogens) need to be studied in future studies. Carman et al. (2017) reported that the mortality of spider crabs increased with Gonionemus consumption, and 100% of the spider crabs died within 24 h after consuming jellyfish with toxic effects, suggesting that Gonionemus sp. medusae can feed on hard-bodied organisms such as copepods and cladocerans as well (Govindarajan et al., 2019). It is notorious for causing severe stings in humans and is considered invasive or cryptogenic elsewhere. In general, we speculate that the toxicity of hydromedusa G. vertens on sea cucumbers was probably caused by the nematocysts more than pathogenic bacterial infections.
5 CONCLUSIONJellyfish-associated bacterial communities were less diverse than both ambient seawater environments (sea cucumber mariculture area and natural environment), and their compositions were distinct from bacterioplankton communities in seawater. Rhodobacterales, Burkholderiales, Campylobacterales, Rhizobiales, and Oceanospirllales were abundant in the bacterial communities associated with G. vertens in Dongying. In contrast, bacterial communities from Yantai and Dalian were dominated by Kiloniellales, which is related to denitrification and biosynthesis of secondary metabolites. The associated bacterial communities had pronounced differences in different geographical environments; and spatial separation and environmental effects were two critical factors. In addition, pathogen sequencing was not detected in the holothurian mariculture system of the symbiosis of jellyfish G. vertens. Hydromedusa G. vertens might had a great toxicity impact on sea cucumbers than that of the pathogen bacterial infections. Further studies need to reveal more details of pathogenic bacterial in G. vertens and other jellyfish species concerned.
6 DATA AVAILABILITY STATEMENTThe data that support the findings of this study are available from the corresponding author upon request.
Bakker C. 1980. On the distribution of 'Gonionemus vertens'A.Agassiz (Hydrozoa, Limnomedusae), a new species in the eelgrass beds of Lake Grevelingen (S.W.Netherlands). Hydrobiological Bulletin, 14(3): 186-195.
DOI:10.1007/BF02260120 |
Basso L, Rizzo L, Marzano M, Intranuovo M, Fosso B, Pesole G, Piraino S, Stabili L. 2019. Jellyfish summer outbreaks as bacterial vectors and potential hazards for marine animals and humans health? The case of Rhizostoma pulmo (Scyphozoa, Cnidaria). Science of the Total Environment, 692: 305-318.
DOI:10.1016/j.scitotenv.2019.07.155 |
Baxter E J, McAllen R, Allcock A L, Doyle T K. 2012. Abundance, distribution and community composition of small gelatinous zooplankton in southern Irish coastal waters. Biology and Environment: Proceedings of the Royal Irish Academy, 112B(1): 91-103.
DOI:10.1353/bae.2012.0029 |
Bernasconi R, Stat M, Koenders A, Huggett M J. 2019a. Global networks of Symbiodinium-bacteria within the coral holobiont. Microbial Ecology, 77(3): 794-807.
DOI:10.1007/s00248-018-1255-4 |
Bernasconi R, Stat M, Koenders A, Paparini A, Bunce M, Huggett M J. 2019b. Establishment of coral-bacteria symbioses reveal changes in the core bacterial community with host ontogeny. Frontiers in Microbiology, 10: 1529.
DOI:10.3389/fmicb.2019.01529 |
Bokulich N A, Kaehler B D, Rideout J R, Dillon M, Bolyen E, Knight R, Huttley G A, Caporaso J G. 2018. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2's q2-feature-classifier plugin. Microbiome, 6(1): 90.
DOI:10.1186/s40168-018-0470-z |
Bosch-Belmar M, Milisenda G, Girons A, Taurisano V, Accoroni S, Totti C, Piraino S, Fuentes V. 2017. Consequences of stinging plankton blooms on finfish mariculture in the Mediterranean Sea. Frontiers in Marine Science, 4: 240.
DOI:10.3389/fmars.2017.00240 |
Byler K A, Carmi-Veal M, Fine M, Goulet T L. 2013. Multiple symbiont acquisition strategies as an adaptive mechanism in the coral Stylophora pistillata. PLoS One, 8(3): e59596.
DOI:10.1371/journal.pone.0059596 |
Carman M R, Grunden D W, Govindarajan A F. 2017. Species-specific crab predation on the hydrozoan clinging jellyfish Gonionemus sp. (Cnidaria, Hydrozoa), subsequent crab mortality, and possible ecological consequences. PeerJ, 5: e3966.
DOI:10.7717/peerj.3966 |
Cleary D F R, Becking L E, De Voogd N J, Pires A C C, Polónia A R M, Egas C, Gomes N C M. 2013. Habitat- and hostrelated variation in sponge bacterial symbiont communities in Indonesian waters. FEMS Microbiology Ecology, 85(3): 465-482.
DOI:10.1111/1574-6941.12135 |
Cleary D F R, Becking L E, Polónia A R M, Freitas R M, Gomes N C M. 2016. Jellyfish-associated bacterial communities and bacterioplankton in Indonesian Marine lakes. FEMS Microbiology Ecology, 92(5): fiw064.
|
Clinton M, Kintner A H, Delannoy C M J, Brierley A S, Ferrier D E K. 2020. Molecular identification of potential aquaculture pathogens adherent to cnidarian zooplankton. Aquaculture, 518: 734801.
DOI:10.1016/j.aquaculture.2019.734801 |
Cortés-Lara S, Urdiain M, Mora-Ruiz M, Prieto L, Rosselló-Móra R. 2015. Prokaryotic microbiota in the digestive cavity of the jellyfish Cotylorhiza tuberculata. Systematic and Applied Microbiology, 38(7): 494-500.
DOI:10.1016/j.syapm.2015.07.001 |
Daley M C, Urban-Rich J, Moisander P H. 2016. Bacterial associations with the hydromedusa Nemopsis bachei and scyphomedusa Aurelia aurita from the North Atlantic Ocean. Marine Biology Research, 12(10): 1088-1100.
DOI:10.1080/17451000.2016.1228974 |
Daniels C, Breitbart M. 2012. Bacterial communities associated with the ctenophores Mnemiopsis leidyi and Beroe ovate. FEMS Microbiology Ecology, 82(1): 90-101.
DOI:10.1111/j.1574-6941.2012.01409.x |
Delannoy C M J, Houghton J D R, Fleming N E C, Ferguson H W. 2011. Mauve stingers (Pelagia noctiluca) as carriers of the bacterial fish pathogen Tenacibaculum maritimum. Aquaculture, 311(1-4): 255-257.
DOI:10.1016/j.aquaculture.2010.11.033 |
Ferguson H W, Christian M J D, Hay S, Nicolson J, Sutherland D, Crumlish M. 2010. Jellyfish as vectors of bacterial disease for farmed salmon (Salmo salar). Journal of Veterinary Diagnostic Investigation, 22(3): 376-382.
DOI:10.1177/104063871002200305 |
Fodelianakis S, Papageorgiou N, Pitta P, Kasapidis P, Karakassis I, Ladoukakis E D. 2014. The pattern of change in the abundances of specific bacterioplankton groups is consistent across different nutrient-enriched habitats in Crete. Applied and Environmental Microbiology, 80(13): 3784-3792.
DOI:10.1128/AEM.00088-14 |
Fringuelli E, Savage P D, Gordon A, Baxter E J, Rodger H D, Graham D A. 2012. Development of a quantitative realtime PCR for the detection of Tenacibaculum maritimum and its application to field samples. Journal of Fish Diseases, 35(8): 579-590.
DOI:10.1111/j.1365-2761.2012.01377.x |
Gallardo B, Clavero M, Sánchez M I, Vilà M. 2016. Global ecological impacts of invasive species in aquatic ecosystems. Global Change Biology, 22(1): 151-163.
DOI:10.1111/gcb.13004 |
Gaynor J J, Bologna P A X, Restaino D, Barry C L. 2016. First occurrence of the invasive hydrozoan Gonionemus vertens A.Agassiz, 1862 (Cnidaria: Hydrozoa) in New Jersey, USA. BioInvasions Records, 5(4): 233-237.
DOI:10.3391/bir.2016.5.4.07 |
Gerpe D, Buján N, Diéguez A L, Lasa A, Romalde J L. 2017. Kiloniella majae sp.nov., isolated from spider crab (Maja brachydactyla) and pullet carpet shell clam (Venerupis pullastra). Systematic and Applied Microbiology, 40(5): 274-279.
DOI:10.1016/j.syapm.2017.05.002 |
Gong L X, Wang H N, Wang T X, Liu Y L, Wang J, Sun B G. 2019. Feruloylated oligosaccharides modulate the gut microbiota in vitro via the combined actions of oligosaccharides and ferulic acid. Journal of Functional Foods, 60: 103453.
DOI:10.1016/j.jff.2019.103453 |
Govindarajan A F, Carman M R, Khaidarov M R, Semenchenko A, Wares J P. 2017. Mitochondrial diversity in Gonionemus(Trachylina: Hydrozoa) and its implications for understanding the origins of clinging jellyfish in the Northwest Atlantic Ocean. PeerJ, 5: e3205.
DOI:10.7717/peerj.3205 |
Govindarajan A F, Carman M R. 2016. Possible cryptic invasion of the Western Pacific toxic population of the hydromedusa Gonionemus vertens (Cnidaria: Hydrozoa) in the Northwestern Atlantic Ocean. Biological Invasions, 18(2): 463-469.
DOI:10.1007/s10530-015-1019-8 |
Govindarajan A F, Källström B, Selander E, Östman C, Dahlgren T G. 2019. The highly toxic and cryptogenic clinging jellyfish Gonionemus sp.(Hydrozoa, Limnomedusae) on the Swedish west coast. PeerJ, 7(e6883).
|
Hao W J, Gerdts G, Peplies J, Wichels A. 2015. Bacterial communities associated with four ctenophore genera from the German Bight (North Sea). FEMS Microbiology Ecology, 91(1): 1-11.
|
Jang H, Yang S H, Seo H S, Lee J H, Kim S J, Kwon K K. 2015. Amphritea spongicola sp.nov., isolated from a marine sponge, and emended description of the genus Amphritea. International Journal of Systematic and Evolutionary Microbiology, 65(Pt6): 1866-1870.
|
Jaspers C, Weiland-Bräuer N, Rühlemann M C, Baines J F, Schmitz R A, Reusch T B H. 2020. Differences in the microbiota of native and non-indigenous gelatinous zooplankton organisms in a low saline environment. Science of the Total Environment, 734: 139471.
DOI:10.1016/j.scitotenv.2020.139471 |
Langille M G I, Zaneveld J, Caporaso J G, Mcdonald D, Knights D, Reyes J A, Clemente J C, Burkepile D E, Thurber R L V, Knight R, Beiko R G, Huttenhower C. 2013. Predictive functional profiling of microbial communities using 16s RRNA marker gene sequences. Nature Biotechnology, 31(9): 814-821.
DOI:10.1038/nbt.2676 |
Lee M D, Kling J D, Araya R, Ceh J. 2018. Jellyfish life stages shape associated microbial communities, while a core microbiome is maintained across all. Frontiers in Microbiology, 9: 1534.
DOI:10.3389/fmicb.2018.01534 |
Lee O O, Wang Y, Yang J K, Lafi F F, Al-Suwailem A, Qian P Y. 2011. Pyrosequencing reveals highly diverse and species-specific microbial communities in sponges from the Red Sea. The ISME Journal, 5(4): 650-664.
DOI:10.1038/ismej.2010.165 |
Lindh M V, Riemann L, Baltar F, Romero-Oliva C, Salomon P S, Granéli E, Pinhassi J. 2013. Consequences of increased temperature and acidification on bacterioplankton community composition during a mesocosm spring bloom in the Baltic Sea. Environmental Microbiology Reports, 5(2): 252-262.
DOI:10.1111/1758-2229.12009 |
Marchessaux G, Gadreaud J, Martin-Garin B, Thiéry A, Ourgaud M, Belloni B, Thibault D. 2017. First report of the invasive jellyfish Gonionemus vertens A.Agassiz, 1862 in the Berre Lagoon, southeast France. Bioinvasions Records, 6(4): 339-344.
DOI:10.3391/bir.2017.6.4.06 |
Mariottini G L, Giacco E, Pane L. 2008. The mauve stinger Pelagia noctiluca (Forsskål, 1775).Distribution, ecology, toxicity and epidemiology of stings. Marine Drugs, 6(3): 496-513.
|
Martiny J B H, Bohannan B J M, Brown J H, Colwell R K, Fuhrman J A, Green J L, Horner-Devine M C, Kane M, Krumins J A, Kuske C R, Morin P J, Naeem S, Øvreås L, Reysenbach A L, Smith V H, Staley J T. 2006. Microbial biogeography: putting microorganisms on the map. Nature Reviews Microbiology, 4(2): 102-112.
DOI:10.1038/nrmicro1341 |
Pyšek P, Richardson D M. 2010. Invasive species, environmental change and management, and health. Annual Review of Environment and Resources, 35: 25-55.
DOI:10.1146/annurev-environ-033009-095548 |
Rodriguez C S, Pujol M G, Mianzan H W, Genzano G N. 2014. First record of the invasive stinging medusa Gonionemus vertens in the southern hemisphere (Mar del Plata, Argentina). Latin American Journal of Aquatic Research, 42(3): 653-657.
DOI:10.3856/vol42-issue3-fulltext-23 |
Rohwer F, Seguritan V, Azam F, Knowlton N. 2002. Diversity and distribution of coral-associated bacteria. Marine Ecology Progress Series, 243: 1-10.
DOI:10.3354/meps243001 |
Ruiz G M, Fofonoff P W, Carlton J T, Wonham M J, Hines A H. 2000. Invasion of coastal marine communities in North America: apparent patterns, processes, and biases. Annual Review of Ecology and Systematics, 31: 481-531.
DOI:10.1146/annurev.ecolsys.31.1.481 |
Russell S L, Corbett-Detig R B, Cavanaugh C M. 2017. Mixed transmission modes and dynamic genome evolution in an obligate animal-bacterial symbiosis. The ISME Journal, 11(6): 1359-1371.
DOI:10.1038/ismej.2017.10 |
Schuchert P. 2016. Gonionemus vertens A. Agassiz, 1862. In: Schuchert P ed. World Hydrozoa Database. Accessed through: World Register of Marine Species at http://www.marinespecies.org/phia.php?p=taxdetails&id=117768 on 2016-10-25.
|
Schuett C, Doepke H. 2010. Endobiotic bacteria and their pathogenic potential in cnidarian tentacles. Helgoland Marine Research, 64(3): 205-212.
DOI:10.1007/s10152-009-0179-2 |
Si O J, Yang H Y, Hwang C Y, Kim S J, Choi S B, Kim J G, Jung M Y, Kim S G, Roh S W, Rhee S K. 2017. Kiloniella antarctica sp.nov., isolated from a polynya of Amundsen Sea in Western Antarctic Sea. International Journal of Systematic and Evolutionary Microbiology, 67(7): 2397-2402.
DOI:10.1099/ijsem.0.001968 |
Småge S B, Brevik Ø J, Frisch K, Watanabe K, Duesund H, Nylund A. 2018. Concurrent jellyfish blooms and tenacibaculosis outbreaks in Northern Norwegian Atlantic salmon (Salmo salar) farms. PLoS One, 12(11): e0187476.
|
Taylor M W, Radax R, Steger D, Wagner M. 2007. Spongeassociated microorganisms: evolution, ecology, and biotechnological potential. Microbiology and Molecular Biology Reviews, 71(2): 295-347.
DOI:10.1128/MMBR.00040-06 |
Tinta T, Kogovšek T, Klun K, Malej A, Herndl G J, Turk V. 2019. Jellyfish-associated microbiome in the marine environment: exploring its biotechnological potential. Marine Drugs, 17(2): 94.
DOI:10.3390/md17020094 |
Toranzo A E, Magariños B, Romalde J L. 2005. A review of the main bacterial fish diseases in mariculture systems. Aquaculture, 246(1-4): 37-61.
DOI:10.1016/j.aquaculture.2005.01.002 |
Viver T, Orellana L H, Hatt J K, Urdiain M, Díaz S, Richter M, Antón J, Avian M, Amann R, Konstantinidis K T, Rosselló-Móra R. 2017. The low diverse gastric microbiome of the jellyfish Cotylorhiza tuberculata is dominated by four novel taxa. Environmental Microbiology, 19(8): 3039-3058.
DOI:10.1111/1462-2920.13763 |
Wahl M, Goecke F, Labes A, Dobretsov S, Weinberger F. 2012. The Second skin: ecological role of epibiotic biofilms on marine organisms. Frontiers in Microbiology, 3: 292.
|
Walsh E A, Kirkpatrick J B, Rutherford S D, Smith D C, Sogin M, D'Hondt S. 2016. Bacterial diversity and community composition from seasurface to subseafloor. The ISME Journal, 10(4): 979-989.
DOI:10.1038/ismej.2015.175 |
Wang K, Ye X S, Chen H P, Zhao Q F, Hu C J, He J Y, Qian Y X, Xiong J B, Zhu J L, Zhang D M. 2015a. Bacterial biogeography in the coastal waters of northern Zhejiang, East China Sea is highly controlled by spatially structured environmental gradients. Environmental Microbiology, 17(10): 3898-3913.
DOI:10.1111/1462-2920.12884 |
Wang L P, Li X Y, Shao Z Z. 2015b. Draft genome sequence of the denitrifying strain Kiloniella sp.P1-1 isolated from the gut microflora of Pacific white shrimp, Litopenaeus vannamei. Marine Genomics, 24: 261-263.
DOI:10.1016/j.margen.2015.07.015 |
Webster G, O'Sullivan L A, Meng Y Y, Williams A S, Sass A M, Watkins A J, Parkes R J, Weightman A J. 2015. Archaeal community diversity and abundance changes along a natural salinity gradient in estuarine sediments. FEMS Microbiology Ecology, 91(2): 1-18.
|
Webster N S, Taylor M W, Behnam F, Lücker S, Rattei T, Whalan S, Horn M, Wagner M. 2010. Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts. Environmental Microbiology, 12(8): 2070-2082.
DOI:10.1111/j.1462-2920.2009.02065.x |
Wei G S, Li M C, Li F E, Li H, Gao Z. 2016. Distinct distribution patterns of prokaryotes between sediment and water in the Yellow River estuary. Applied Microbiology and Biotechnology, 100(22): 9683-9697.
DOI:10.1007/s00253-016-7802-3 |
Weiland-Bräuer N, Neulinger S C, Pinnow N, Künzel N, Künzel S, Baines J F, Schmitz R A. 2015. Composition of bacterial communities associated with Aurelia aurita changes with compartment, life stage, and population. Applied and Environmental Microbiology, 81(17): 6038-6052.
DOI:10.1128/AEM.01601-15 |
Wiese J, Thiel V, Gärtner A, Schmaljohann R, Imhoff J F. 2009. Kiloniella laminariae gen.nov., sp.nov., an alphaproteobacterium from the marine macroalga Laminaria saccharina. International Journal of Systematic and Evolutionary Microbiology, 59(Pt 2): 350-356.
|
 2022, Vol. 40
2022, Vol. 40


