Institute of Oceanology, Chinese Academy of Sciences
Article Information
- FANG Kunpeng, NAN Fangru, FENG Jia, LÜ Junping, LIU Qi, LIU Xudong, XIE Shulian
- Pauciramus yunnanensis, gen. et sp. nov., a novel freshwater red alga from China with proposal of the Ottiales ord. nov. (Nemaliophycidae, Rhodophyta)
- Journal of Oceanology and Limnology, 40(3): 1245-1256
- http://dx.doi.org/10.1007/s00343-021-1154-2
Article History
- Received May. 7, 2021
- accepted in principle Jun. 28, 2021
- accepted for publication Aug. 16, 2021
Red algae (Rhodophyta) primarily inhabit marine environments, with freshwater red algae accounting for only 3% of the species diversity (Sheath and Vis, 2015). These freshwater taxa are particularly well represented in the subclass Nemaliophycidae, including four orders: Acrochaetiales, Balbianiales, Batrachospermales, and Thoreales (Kumano, 2002; Lam et al., 2016). The order Acrochaetiales currently includes four families with 220 accepted species and is the only order of subclass Nemaliophycidae that has both freshwater and marine taxa, with approximately 10% of species occurring in freshwater environment (Guiry and Guiry, 2021). The freshwater taxa are only distributed in the genera Audouinella Bory and Ottia Entwisle, J. R. Evans, M. L. Vis, & G. W. Saunders (Harper and Saunders, 2002; Kumano, 2002; Entwisle et al., 2018). Ottia belongs to the monogeneric family Ottiaceae with only one species Ottia meiospora (Skuja) Entwisle, J. R. Evans, M. L. Vis, & G. W. Saunders. This genus and family were recently established based on morphological characteristics and DNA sequence data; this taxon is an endophyte of other freshwater rhodophytes and characterized by uniseriate branched filaments, cells with 1-2 parietal, cup, or ribbon-shaped chloroplasts, without pyrenoids, and monosporangia in small clusters (Entwisle et al., 2018). Skuja (1944) first described O. meiospora (as Balbiania meiospora Skuja) from a collection of Nothocladus lindaueri Skuja from the Bay of Islands, New Zealand. Garbary (1987) transferred Balbiania (only includes two species B. meiospora and B. investiens (Lenormand ex Kützing) Sirodot) to Audouinella due to no obvious morphological difference between Balbiania and Audouinella. Subsequently, based on molecular comparisons and further study of morphology, B. investiens was returned to the genus Balbiania with a new family (Balbianiaceae) and order (Balbianiales) established. By contrast, Audouinella (Balbiania) meiospora remained tentatively assigned to Audouinella (family Audouinellaceae) due the lack of DNA sequence data (Harper and Saunders, 1998; Sheath and Müller, 1999). Soon after, Skinner and Entwisle (2001) identified an Australian endophyte of Nothocladus from northern New South Wales and Little Yarra River in Victoria and attributed collection to B. investiens. Entwisle et al. (2018) reinvestigated this Australian taxon. Their phylogenetic analyses confirmed that it belonged in the order Acrochaetiales, and Australian collections have a similar morphology B. meiospora as reported by Skuja (1944). To accommodate this red algal endophyte, a new genus and family were proposed (Entwisle et al., 2018).
Freshwater taxa of Acrochaetiales are widely distributed in China. Since Jao (1940) first reported Acrochaetiales species in freshwater environments from China, 10 species have been reported, all of which belong to the genus Audouinella (Shi, 2006). However, four have been assigned to other taxa or the sporophyte stages of other freshwater taxa using molecular analyses (Pueschel et al., 2000; Chiasson et al., 2005, 2007; Necchi and Oliveira, 2011; Chen et al., 2014), and the remaining six species are still to be investigated with molecular data to support their distinctiveness. Xie and Shi (2004) reported B. meiospora, endophyte of batrachspermalean taxa, using morphology from a collection of Zhejiang (Hangzhou, Yuhuang Mountain, CK547, collected by C.-C. Jao in 1954) and Guizhou (Huangguoshu, KC830108, collected by Yong YAO et al. in 1983). Whether this Chinese collection is Ottia meiospora requires further confirmation. In this study, we discovered an interesting uniseriate freshwater red alga from Ailao Mountain, Yunnan (Fig. 1). Phylogenetic analyses demonstrate its position within the family Ottiaceae clade and requires a new systematic arrangement with the establishment of a new genus and order.
|
| Fig.1 Map of China showing the location of the study area Ailao Mountain in Yunnan, China Map review No. GS(2020)4634. |
Algal specimens were collected in April 2019 from Shimenxia Scenic Area of Ailao Mountain, Yunnan, China (23°58'22"N, 101°31'25"E, above sea level (ASL): 1 987.7 m Fig. 1). The clear stream flowing from the valley of Ailao Mountain passes through nearly 1 km of rocky canyons (Shimenxia Scenic Area) surrounded by dense forests. The spring water formed many small streams, which flowed down from the cliffs of the canyon and merged into the river. The algae only grow on the wet and smooth rocks, hit by drops of mountain spring water, on the banks of the river.
The samples were washed with sterile water to remove debris and epiphytes. Specimens for DNA analysis were preserved in silica desiccant. Some voucher specimens were preserved in 4% formaldehyde (holotype: SXU-YN19053) for morphological analyses and the remaining portions were pressed as herbarium voucher specimens (isotype: SXU-YN19054, SXU-YN19055). Voucher specimens were deposited in Herbarium of Shanxi University (SXU), Shanxi University, Taiyuan, Shanxi Province, China. Herbarium acronym follows Thiers (2021).
2.2 Morphological and DNA analysesMorphological analyses were conducted according to previous literature (Skuja, 1944; Skinner and Entwisle, 2001; Kumano, 2002; Xie and Shi, 2004; Shi, 2006; Entwisle et al., 2018). Vegetative and reproductive structures were photographed and examined using an Olympus BX-43 microscope equipped with a charge-coupled device (DP72; Olympus, Tokyo, Japan).
For DNA extraction, tissue dried in silica was homogenized by grinding in liquid nitrogen with a mortar and pestle, and total DNA was extracted following the protocol described by Saunders (1993) with modifications following Vis and Sheath (1997). Polymerase chain reaction (PCR) amplification of the plastid-encoded ribulose-1, 5-bisphosphate carboxylase-oxygenase large-subunit gene (rbcL), barcode region near the 5' end of the mitochondrial cytochrome oxidase subunit I gene (COI-5P) and small subunit gene of the ribosomal cistron (SSU) were chosen for amplification in 20-μL volumes containing 11.3-μL ddH2O, 2-μL 10×buffer, 2.0-μL 2.5-mmol/L dNTPs, 0.2-μL Taq DNA polymerase (all from Sangon Biotech Co., Ltd., China), 2.0 μL of each primer (10 mmol/L), and 0.5 μL of genomic DNA. The rbcL gene (1 532 bp) was PCR amplified using the F-57×R-753, F-993×R-rbcLS primers (Freshwater and Rueness, 1994) and F160×rbcLR primers (Vis et al., 1998) using the following cycle: 95 ℃ for 3 min, 40 cycles of 94 ℃ for 1.5 min, 37 ℃ for 2 min, 72 ℃ for 3 min, a final hold for 2 min at 72 ℃; the COI-5P gene (664 bp) was PCR amplified using the GazF1 and GazR1 primers (Saunders, 2005) using the following cycle: 94 ℃ for 2 min, 35 cycles of 94 ℃ for 30 s, 47 ℃ for 30 s, 72 ℃ for 1 min, a final hold for 7 min at 72 ℃; and the SSU gene (1 572 bp) was PCR amplified using G01×G10, G02×G14, G04×G13, and G06×G17 primers (Harper and Saunders, 2001) using the following cycle: 94 ℃ for 4 min, 38 cycles of 94 ℃ for 30 s, 55 ℃ for 30 s, 72 ℃ for 1 min 30 s, a final hold for 7 min at 72 ℃.
2.3 Sequence determination and data analysisThe PCR products were purified using a SanPrep column DNA gel purification kit (Sangon, China) and then were submitted to BGI Tech Corporation (Beijing, China) for sequencing using an ABI 3730XL sequencer with PCR primers. Contigs were assembled and edited using Sequencher 4.10.1. The sequence data of rbcL (MW874643), COI-5P (MW874644) and SSU (MW879164) markers were deposited in GenBank.
For phylogenetic analyses, sequence data for subclass Nemaliophycidae were downloaded from GenBank (Supplementary Table S1 for analysis based on rbcL, Supplementary Table S2 for analysis based on rbcL+SSU+COI-5P). The subclass Corallinophycidae was used as outgroup for phylogenetic analyses (Scott et al., 2013).
Four alignments were constructed using BioEdit v7.2.1 (Hall, 1999) with default gap penalties and refined by eye, including individual gene regions rbcL (rbcL1: 87 from 53 species, 1 282 aligned sites for analysis based on rbcL) (rbcL2: 56 from 53 species, 1 282 aligned sites for analysis based on rbcL+SSU+COI-5P), SSU (51 from 50 species, 1 791 aligned) and COI-5P (44 from 44 species, 664 aligned). In addition, a combined alignment was generated (57 taxa, 3 737 aligned sites; rbcL=1-1 282, SSU=1 283-3 073, and COI-5P=3 074-3 737) using PhyloSuite (Zhang et al., 2020). Pairwise distance comparison of sequence variation was conducted using MEGA5 software (Supplementary Table S3) (Tamura et al., 2011), and Box-plot of genetic distance based on rbcL, SSU, and COI-5P gene sequences was constructed using OriginPro 2021 (OriginLab Corporation, Northampton, MA, USA). The best-fit model for rbcL were determined by ModelFinder (Kalyaanamoorthy et al., 2017) with AIC criterion (GTR+F+R5 for maximum likelihood (ML) and GTR+F+I+G4 for Bayesian Inference (BI)); and for the concatenated rbcL+SSU+COI-5P alignment, the appropriate models of sequence evolution were selected using PartitionFinder2 (Lanfear et al., 2016), with all algorithm and AIC criterion (for ML: Subset (1)(2)(3)=GTR+I+G; for BI, lset applyto (1)(2)(3): nst=6, rates=invgamma). ML phylogenies were inferred using IQ-TREE (Nguyen et al., 2015) with 5 000 ultrafast bootstraps (for rbcL, under the GTR+R5+F model and for rbcL+SSU+COI-5P under Edge-linked partition model), as well as the Shimodaira-Hasegawa-like approximate likelihood-ratio test (Guindon et al., 2010). Bayesian Inference phylogenies were inferred using MrBayes 3.2.6 (Ronquist et al., 2012) with 2 parallel runs, 2 000 000 generations, in which the initial 25% of sampled data were discarded as burn-in (for rbcL under GTR+I+G+F model; and for rbcL+SSU+COI-5P under partition model). The resulting phylogenetic trees were edited using Figtree1.4.2 (https://tree.bio.ed.ac.uk/software/figtree/).
3 RESULT 3.1 Molecular analysisThree sequences (rbcL-1 282 bp, COI-5P-664 bp, SSU-1 791 bp) were obtained from the samples SXU-YN19053. Unfortunately, despite the best efforts, we were unable to grow this alga in culture, and the sequencing of LSU (large subunit gene of the ribosomal cistron) failed with repeated attempts using a variety of standard primer sets and protocols. Four alignments of the subclass Nemaliophycidae were generated. The rbcL1 data set (for analysis based on rbcL) is 1 282 base pairs with 658 constant sites and 624 variable sites; The rbcL2 data set (for analysis based on rbcL+SSU+COI-5P) is 1 282 base pairs with 670 constant sites and 612 variable sites; the COI-5P data consisted of 664 characters, of which 296 are constant sites and 368 are variable sites; and alignment of SSU gene is 1 791 bp, which included 1 134 constant sites and 654 variable sites.
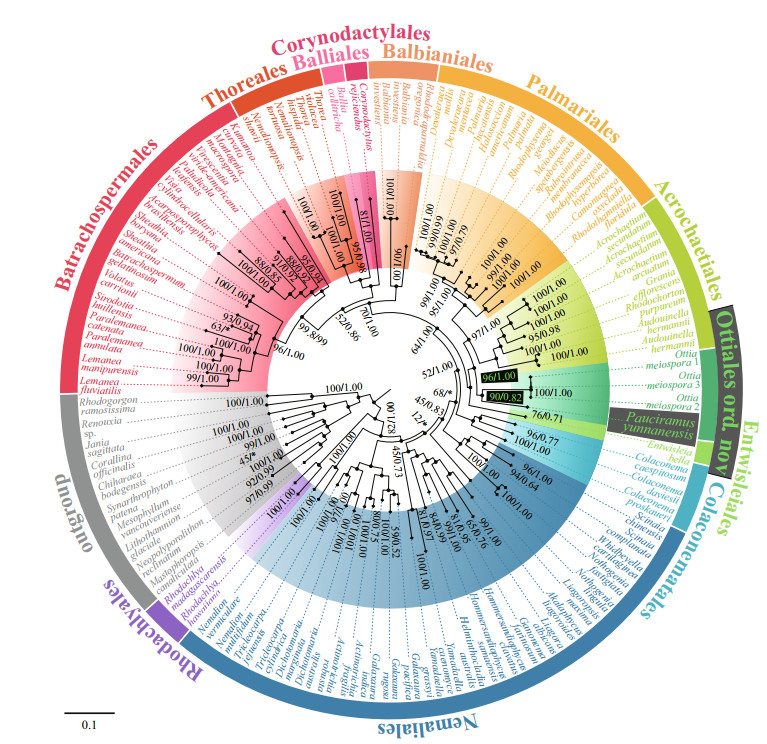
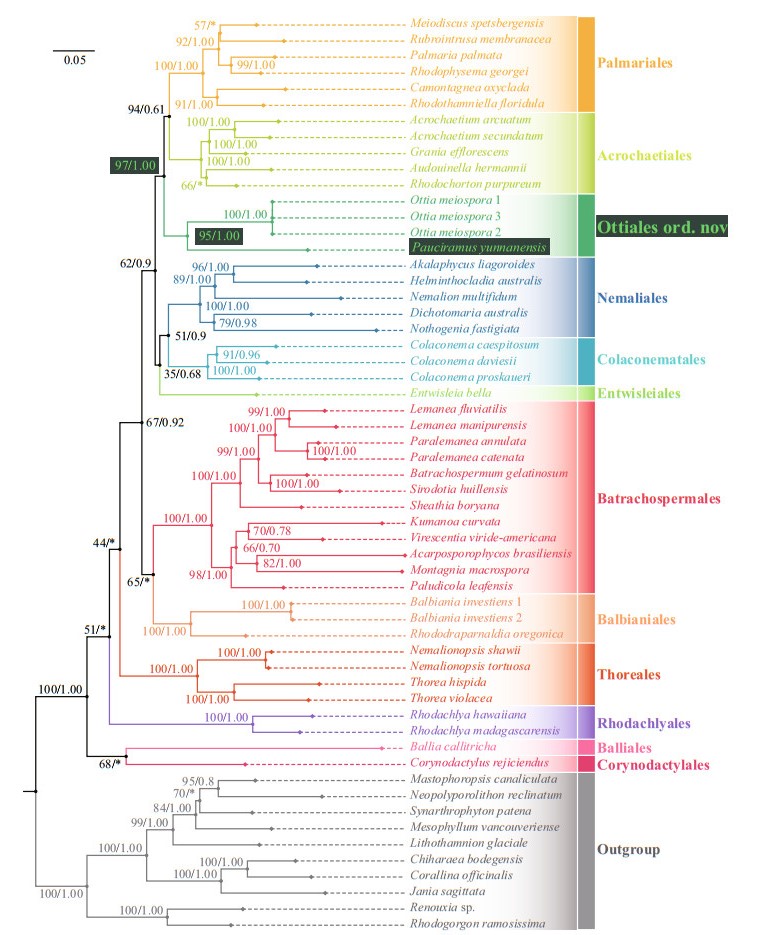
The phylogenetic trees generated from ML and BI analyses based on rbcL or the concatenated rbcL+SSU+COI-5P alignment were closely congruent, such that only the trees inferred from ML were presented here with ML bootstrap support values and BI posterior probability on the nodes (Figs. 2 & 3). Both trees showed the Ailao Mountain specimen was in a well-supported clade with Ottia meiospora (90/0.82 for rbcL tree and 95/1.00 for rbcL+SSU+COI-5P tree) with high interspecific divergences (13.40%, 163 bp for rbcL, and 0% among the three samples of O. meiospora, O. meiospora only have rbcL and LSU sequences), and sister to the order Palmariales and other families of the Acrochaetiales (96/1.00 for rbcL tree and 97/1.00 for rbcL+SSU+COI-5P tree), making the order Acrochaetiales polyphyletic. The great genetic distance between SXU-YN19053 and O. meiospora indicate that the specimen in this study represent distinct species and genus, and Pauciramus yunnanensis is proposed.
|
| Fig.2 Maximum likelihood (ML) phylogeny of subclass Nemaliophycidae estimated from a concatenated alignment of the rbcL Different colors represent different orders. Numbers at the branches indicate the bootstrap (BP, left) support for ML and posterior probabilities (PP, right) for Bayesian inference (BI). "*" denotes the branch differed in the BI topology. Specimen from this study is indicated in black box. |
|
| Fig.3 Maximum likelihood (ML) phylogeny of subclass Nemaliophycidae estimated from a concatenated alignment of the rbcL+SSU+COI-5P markers Different colors represent different order. Numbers at the branches indicate the bootstrap (BP, left) support for ML and posterior probabilities (PP, right) for Bayesian inference (BI). "*" denotes the branch differed in the BI topology. Specimen from this study is indicated in black box. |
To adhere to the principle of monophyly, a new order including the genera Ottia and Pauciramus is proposed. The genetic distance of Acrochaetiales-Palmariales complex also supports this taxonomic decision (Supplementary Figs.S1-S3 and Supplementary Table S3). The genetic distance levels between Ottia-Pauciramus complex and the other taxa of Acrochaetiales-Palmariales complex (including order Acrochaetiales, order Palmariales, and the only freshwater group Audouinella) are significantly higher than the genetic distance between these taxa. This means that Ottia-Pauciramus complex has reached the order level in terms of genetic distance. In addition, we also obtained some interesting results through these genetic distance analyses. The results show that the rbcL sequence has better distinguishability at different taxonomic levels of order — 10.77%-15.21% (131-185 bp), family — 6.09%-11.18% (74-136 bp), and genus — 0.99%-9.21% (12-112 bp), but the genus Audouinella seems to be a special case with generally large genetic distance, almost reaching the order level (10.44%- 14.88%, 127-181 bp) (Supplementary Fig.S1). SSU results (Supplementary Fig.S2) show that Ottia-Pauciramus complex is significantly different from other taxa (3.38%-4.20%, 53-66 bp), and the divergences of taxa in order Palmariales (0.38%- 2.04%, 6-32 bp) is greater than order Acrochaetiales (0.32%-0.64%, 5-10 bp). COI-5P (Supplementary Fig.S3) generally has a large level of difference among all taxa of Palmariales-Acrochaetiales complex (13.03%-23.29%, 80-143 bp), and there is no hierarchical difference related to the classification level.
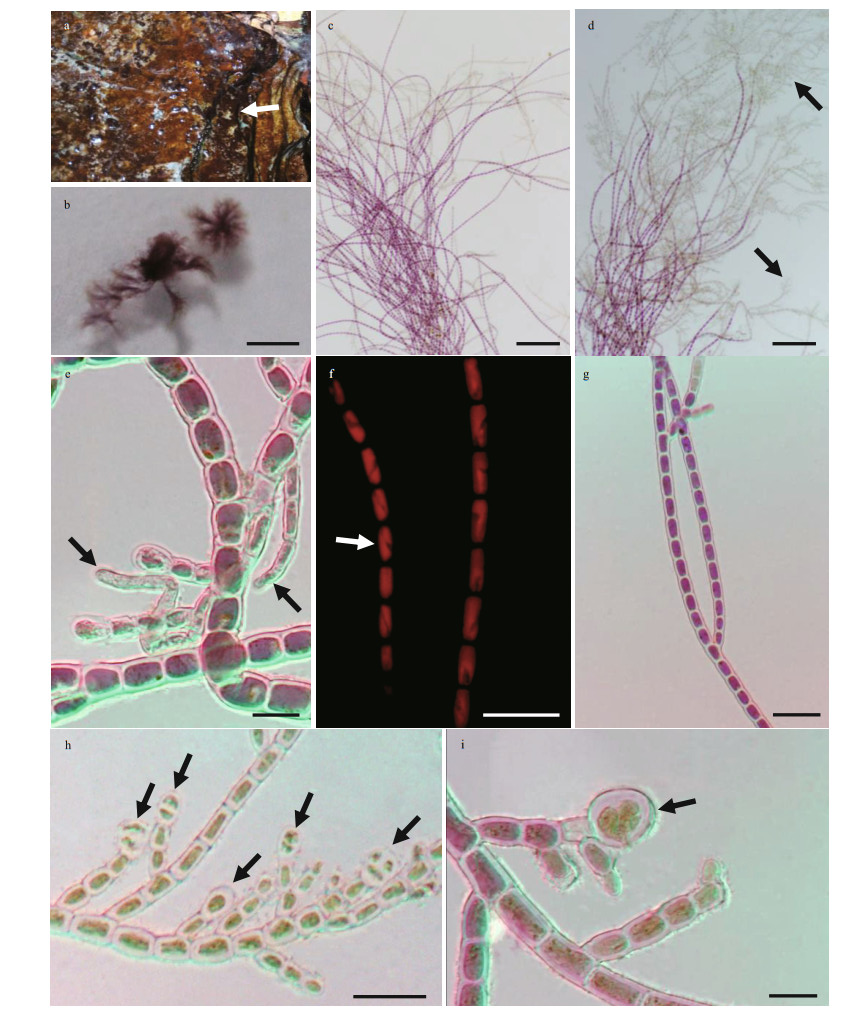
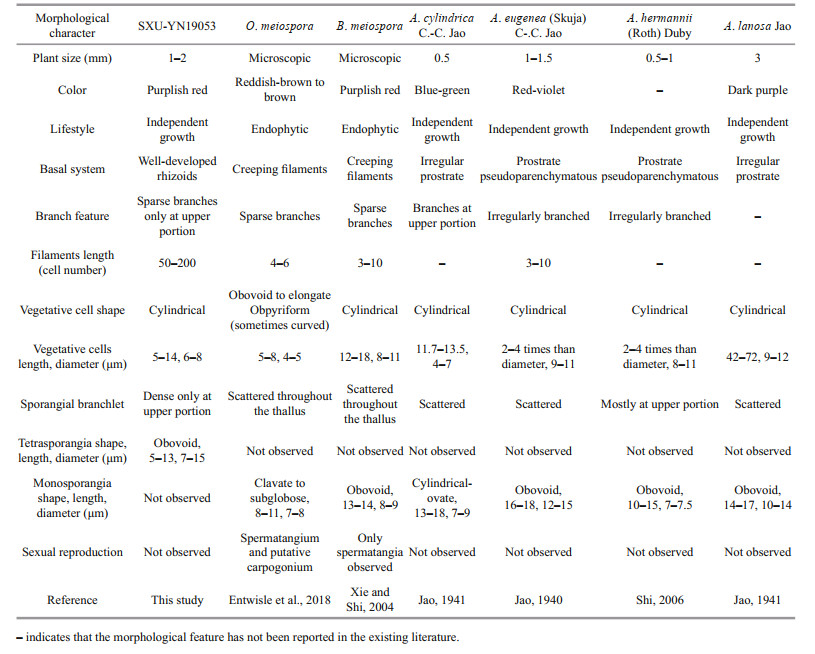
3.2 Morphological observationThe vegetative structures of the specimen (Fig. 4) were caespitose, densely pulvinate, consisting of uniseriate, slender filaments, and attached to the substratum by well-developed rhizoids. These characteristics are close to some taxa of order Acrochaetiales such as Audouinella, Balbiania, and Ottia. Morphological comparison of sample in this study with species of Audouinella and Balbiania previously reported from China and Ottia meiospora is provided (Table 1) according to the descriptions in earlier studies (Skuja, 1944; Kumano, 2002; Xie and Shi, 2004, Shi, 2006; Skinner and Entwisle, 2001; Entwisle et al., 2018).
|
| Fig.4 Morphological structures of Pauciramus yunnanensis, gen. et sp. nov. a. habitat in the wet and smooth rocks on the banks of the river of Ailao Mountain. Densely pulvinate thallus of P. yunnanensis (white arrow); b. vegetative caespitose thalli; c. sparsely branched slender filaments; d. dense sporangial branchlet (arrows) distributed at the upper portion of filaments; e. well-developed rhizoids (arrows) forming from basal cells of filaments; f-g. cylindrical vegetative cells with a single, ribbon-shaped, parietal chloroplast (arrow); h. cruciately divided tetrasporangia (arrows), terminal on the short lateral branchlets; i. cruciately divided tetrasporangium details (arrow). Scale bars: 2 mm in b, 100 μm in c-d, 20 μm in f-h, and 10 μm in e, i. |
|
Our sample (SXU-YN19053) has obvious morphological differences with O. meiospora including slender filaments 50-200 cell stories with well-developed rhizoids (vs. 4-6 cell stories with creeping filaments in O. meiospora); cylindrical vegetative cells (vs. obovoid to elongate obpyriform in O. meiospora); sporangia branchlet only at upper portion (vs. scattered throughout the thallus in O. meiospora); reproduction by tetrasporangia (vs. by monospores in O. meiospora), and the sexual reproduction structure, spermatangium and putative carpogonium reported in O. meiospora was not found in the newly collected freshwater red algae. Furthermore, O. meiospora is endophyte, and epiphytic/endophytic on various species of Nothocladus s. lat., spreading loosely throughout thallus of host (Entwisle et al., 2018), which is obviously different from sample in this study as our samples were caespitose with epilithic habit.
In China, six freshwater Audouinella and one Balbiania have been reported (Xie and Shi, 2004; Shi, 2006), and all of these species lack molecular sequences. However, these species can be easily distinguished from the samples in this study by morphology, especially the well-developed rhizoids and tetrasporangia. Furthermore, B. meiospora previously reported from China can be identified as O. meiospora based on morphology.
3.3 Taxonomic proposalOttiales K. P. Fang, F. R. Nan, & S. L. Xie ord. nov. Diagnosis: Small freshwater red alga composed of uniseriate branched filaments, cells with a single or sometimes two parietal chloroplasts; chloroplast cup or ribbon-shaped, without pyrenoids; fitting one of the two following sets of characteristics: (1) Epiphytic/endophytic on various species of Nothocladus s. lat., consisting of basal and erect systems; cells of basal irregular in shape, fusiform to occasionally inflated in middle or ends; erect systems branched, 4-6-celled, cells obovoid to elongate obpyriform (sometimes curved), terminal hairs present; asexual reproduction by monospores; sexual reproduction with spermatangium and putative carpogonium. (2) Epilithic on rock substrata, consisting of caespitose and densely pulvinate thalli, gracile and uniseriate filaments with well-developed rhizoids, rarely branched; cells cylindrical, terminal hairs absent; asexual reproduction by tetrasporangia, sporangial branchlet only at the upper portion of filaments; sexual reproduction unknown.
Type family: Ottiaceae (Entwisle, J. R. Evans, M. L. Vis, & G. W. Saunders) emend. K. P. Fang, F. R. Nan, & S. L. Xie
Ottiaceae (Entwisle, J. R. Evans, M. L. Vis, & G. W. Saunders) emend. K. P. Fang, F. R. Nan, & S. L. Xie
Characters as for the order.
Type genus: Ottia Entwisle, J. R. Evans, M. L. Vis, & G. W. Saunders (2018: 82)
Taxonomic notes: Family Ottiaceae was established with only one genus and species Ottia meiospora based on morphological characteristics and DNA sequence data (Entwisle et al., 2018). In this study, we found a uniseriate freshwater red alga, and molecular data from the rbcL, SSU and COI-5P indicated that: the new taxon was in a well-supported clade with Ottia meiospora with high interspecific divergences. Additionally, there are obvious morphological differences between Ottia meiospora and the new taxon. Due to low species diversity with few molecular sequences in this lineage, we decided to propose a new species and genus Pauciramus yunnanensis, and provisionally classified Pauciramus into the family Ottiaceae with necessary revisions to the description of this family for the newly added taxon. The revised Ottiaceae is composed of small freshwater red alga and characterized by uniseriate branched filaments, cells with parietal cup or ribbon-shaped chloroplasts, without pyrenoids, including two taxa, one is epiphytic/endophytic (Ottia) and the other is epilithic (Pauciramus).
Pauciramus K. P. Fang, F. R. Nan, & S. L. Xie gen. nov.
Type species: Pauciramus yunnanensis K. P. Fang, F. R. Nan, & S. L. Xie sp. nov
Description: Small freshwater red alga, caespitose and densely pulvinate, consisting of gracile and uniseriate filaments with well-developed rhizoids, rarely branched; cylindrical vegetative cells with a single, ribbon-shaped and parietal chloroplast, without pyrenoids; reproduction by tetraspores, tetrasporangia single, obovoidal, cruciately divided sporangial branchlet only at the upper portion of filaments. Sexual reproduction unknown.
Etymology: From Latin for the "sparse branch". Pauciramus yunnanensis K. P. Fang, F. R. Nan, & S. L. Xie sp. nov. (Fig. 4a-i)
Description: Plants caespitose, woolly, densely pulvinate, purplish red, about 0.1-0.2-cm high, attached to the substratum by well-developed rhizoids, consisting of uniseriate, gracile filaments. Rhizoids forming from basal cells of filaments, comprising 1-3 cells, 30-80-μm long, 3-5 μm in diameter. Main filaments 50-200 cell-storeys, rarely branched at the base and middle, usually have dense sporangial short lateral branchlet at the upper portion. Vegetative cells cylindrical, 6-8 μm in diameter and 5-14-μm long, 1-2 times longer than diameter gradually increases from the base to the upper of main filaments, more or less constricted at the cross walls, walls 1.5-2.5-μm thick, terminal hairs absent; each cell with a single, ribbon-shaped, parietal chloroplast, without pyrenoids. Reproduction by means of tetrasporangia, single, obovoidal, cruciately divided, 5-13 μm in diameter and 7-15-μm long, terminating short lateral branchlets consisting of 1-3 cells, unequally distributed only at the upper portion of thallus. Sexual reproduction unknown.
Diagnosis: Diagnostic DNA sequence: rbcL, SSU, and COI-5P (accession number: MW874643 for rbcL, MW879164 for SSU, and MW874644 for COI-5P).
Type locality: China―Yunnan, Ailao Mountain (23°58'22"N, 101°31'25"E, ASL: 1 987.7 m). On rocks hit by drops of mountain spring water in Shimenxia, a canyon of Ailao Mountain.
Holotype: Liquid immersion specimen (holotype: SXU-YN19053) and herbarium vouchers (isotype: SXU-YN19054, SXU-YN19055).
Etymology: The species epithet refers to the type locality (Yunnan, China).
4 DISCUSSIONMolecular phylogenetic analyses have had a significant impact on subclass Nemaliophycidae with considerable taxonomic changes at the family and order levels: Balbianiales from Acrochaetiales (Sheath and Müller, 1999), Balliales from Ceramiales (Choi et al., 2000), Colaconematales from Acrochaetiales (Harper and Saunders, 2002), and Thoreales from Batrachospermales (Müller et al., 2002). The taxa of order Acrochaetiales have a simple vegetative structure consisting of monosiphonous, branched, heterotrichous, filaments with apical growth, reproduction by monosporangia or tetrasporangia, sexuality by simple carpogonium (Kumano, 2002), such that it has had the most confusing taxonomic histories in red algae, with about 800 species names and twenty generic names described and used (Harper and Saunders, 2002; Guiry and Guiry, 2021). The introduction of molecular phylogenetic analyses has significantly improved the complicated and chaotic situation. However, the relationship among genera and their familial assignments are still uncertain (Saunders et al., 1995; Volovsek et al., 2000; Harper and Saunders, 2002; Clayden and Saunders, 2008, 2014). In this study, we describe a newly collected uniseriate freshwater red algae (SXU-YN19053) from Ailao Mountain, Yunnan, China, based on morphological observations and phylogenetic analysis.
The phylogenetic trees revealed that Acrochaetiales is polyphyletic. The Ailao Mountain specimen (SXU-YN19053) was in a well-supported clade with Ottia meiospora (from Hawaii, USA) and sister to order Palmariales and other families of the Acrochaetiales. And the genetic distance levels between Ottia-Pauciramus complex and the other taxa of Acrochaetiales-Palmariales complex (including order Acrochaetiales, order Palmariales, and the only freshwater group Audouinella) are significantly higher than the genetic distance between these taxa. This indicates that Ottia-Pauciramus complex may have the same status as Acrochaetiales and Palmariales at the order level, and Ottia-Pauciramus complex can be regarded as an order, so that it can solve the polyphyly of Acrochaetiales.
Molecular phylogenetic analyses based on rbcL, SSU, and COI-5P show that the rbcL sequence has better distinguishability at different taxonomic levels of order, family, and genus, which may be used as a basis for classification of genus, family, and order in the future.
The molecular sequences of the freshwater taxa are very rare in Acrochaetiales-Ottiales-Palmariales at present. Audouinella is the only genus with both marine and freshwater representatives. As molecular analyses have shown that many of these freshwater taxa are sporophyte stages of other freshwater orders, (Necchi and Zucchi, 1997; Pueschel et al., 2000; Chiasson et al., 2005, 2007; Necchi and Oliveira, 2011; Chen et al., 2014), there are now comparatively few species of true Audouinella with only one species A. hermannii supported by molecular data. Many scholars believe that the freshwater groups of Audouinella should be classified into a family Audouinellaceae that needs to be resurrected (Harper and Saunders, 2002; Saunders et al., 2018). Similarly, Ottia has only one species supported by molecular sequence. Therefore, there are only three freshwater species supported by molecular evidence including the newly proposed taxa in this study. For a monospecific genus seems impractical for the establishment of a new family, only a new genus Pauciramus was proposed in a new order Ottiales to accommodate the uniseriate red algal described in this study, and provisionally classified this taxon to the family Ottiaceae.
5 DATA AVAILABILITY STATEMENTThe datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Electronic supplementary materialSupplementary material (Supplementary Tables S1-S3 and Supplementary Figs.S1-S3) is available in the online version of this article at https://doi.org/10.1007/s00343-021-1154-2.
Chen L, Feng J, Han X J, Xie S L. 2014. Investigation of a freshwater acrochaetioid alga (Rhodophyta) with molecular and morphological methods. Nordic Journal of Botany, 32(5): 529-535.
DOI:10.1111/njb.00407 |
Chiasson W B, Johanson K G, Sherwood A R, Vis M L. 2007. Phylogenetic affinities of form taxon Chantransia pygmaea (Rhodophyta) specimens from the Hawaiian Islands. Phycologia, 46(3): 257-262.
DOI:10.2216/06-79.1 |
Chiasson W B, Sabo N J, Vis M L. 2005. Affinities of freshwater putative chantransia stages (Rhodophyta) from molecular and morphological data. Phycologia, 44(2): 163-168.
DOI:10.2216/0031-8884(2005)44[163:AOFPCS]2.0.CO;2 |
Choi H G, Kraft G T, Saunders G W. 2000. Nuclear small-subunit rDNA sequences from Ballia spp. (Rhodophyta): proposal of the Balliales ord. nov., Balliaceae fam. nov., Ballia nana sp. nov. and Inkyuleea gen. nov. (Ceramiales). Phycologia, 39(4): 272-287.
DOI:10.2216/i0031-8884-39-4-272.1 |
Clayden S L, Saunders G W. 2008. Resurrecting the red algal genus Grania within the order Acrochaetiales (Florideophyceae, Rhodophyta). European Journal of Phycology, 43(2): 151-160.
DOI:10.1080/09670260801896630 |
Clayden S L, Saunders G W. 2014. A study of two Acrochaetium complexes in Canada with distinction of Rhododrewia gen. nov. (Acrochaetiales, Rhodophyta). Phycologia, 53(3): 221-232.
DOI:10.2216/13-224.1 |
Entwisle T J, Evans J R, Vis M L, Saunders G W. 2018. Ottia meiospora (Ottiaceae, Rhodophyta), a new genus and family endophytic within the thallus of Nothocladus (Batrachospermales, Rhodophyta). Journal of Phycology, 54(1): 79-84.
DOI:10.1111/jpy.12603 |
Freshwater, D W, Rueness, J. 1994. Phylogenetic relationships of some European Gelidium (Gelidiales, Rhodophyta) species, based on rbcL nucleotide sequence analysis. Phycologia, 33(3): 187-194.
DOI:10.2216/i0031-8884-33-3-187.1 |
Garbary D J. 1987. The Acrochaetiaceae (Rhodophyta): an annotated bibliography. Bibliotheca Phycologica, 77: 1-267.
|
Guindon S, Dufayard J F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology, 59(3): 307-321.
DOI:10.1093/sysbio/syq010 |
Guiry M D, Guiry G M. 2021. AlgaeBase. World-wide electronic publication, National University of Ireland, Galway. https://www.algaebase.org. Accessed on Apr. 25, 2021.
|
Hall T A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symposium Series, 41: 95-98.
|
Harper J T, Saunders G W. 2001. The application of sequences of the ribosomal cistron to the systematics and classification of the Florideophyte red algae (Florideophyceae, Rhodophyta). Cahiers De Biologie Marine, 42: 25-38.
|
Harper J T, Saunders G W. 2002. A re-classification of the Acrochaetiales based on molecular and morphological data, and establishment of the Colaconematales ord. nov. (Florideophyceae, Rhodophyta). European Journal of Phycology, 37(3): 463-476.
DOI:10.1017/s0967026202003840 |
Harper J, Saunders G. 1998. A molecular systematic investigation of the Acrochaetiales (Florideophycidae, Rhodophyta) and related taxa based on nuclear small-subunit ribosomal DNA sequence data. European Journal of Phycology, 33(3): 221-229.
DOI:10.1080/09670269810001736723 |
Jao C C. 1940. Studies on the freshwater algae of China. Subaerial and aquatic algae from Nanyoh, Hunan. Sinensia, 11: 241-360.
|
Jao C C. 1941. Studies on the freshwater algae of China. VIII. A preliminary account of the Chinese freshwater Rhodophyceae. Sinensia, 12: 245-289.
|
Kalyaanamoorthy S, Minh B Q, Wong T K F, von Haeseler A, Jermiin L S. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods, 14(6): 587-589.
DOI:10.1038/nmeth.4285 |
Kumano S. 2002. Freshwater Red Algae of the World. Biopress Ltd, Bristol, UK. 375p.
|
Lam D W, Verbruggen H, Saunders G W, Vis M L. 2016. Multigene phylogeny of the red algal subclass Nemaliophycidae. Molecular Phylogenetics & Evolution, 94: 730-736.
DOI:10.1016/j.ympev.2015.10.015 |
Lanfear R, Frandsen P B, Wright A M, Senfeld T, Calcott B. 2016. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution, 34(3): 772-773.
DOI:10.1093/molbev/msw260 |
Müller K M, Sherwood A R, Pueschel C M, Gutell R R, Sheath R G. 2002. A proposal for a new red algal order, the Thoreales. Journal of Phycology, 38(4): 807-820.
DOI:10.1046/j.1529-8817.2002.01055.x |
Necchi O Jr, Oliveira M C. 2011. Phylogenetic affinities of "chantransia" stages in members of the Batrachospermales and Thoreales (Rhodophyta). Journal of Phycology, 47(3): 680-686.
DOI:10.1111/j.1529-8817.2011.00997.x |
Necchi O, Zucchi M R. 1997. Audouinella macrospora (Acrochaetiaceae, Rhodophyta) is the Chantransia stage of Batrachospermum (Batrachospermaceae). Phycologia, 36(3): 220-224.
DOI:10.2216/i0031-8884-36-3-220.1 |
Nguyen L T, Schmidt H A, von Haeseler A, Minh B Q. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology & Evolution, 32(1): 268-274.
DOI:10.1093/molbev/msu300 |
Pueschel C M, Saunders G W, West J A. 2000. Affinities of the freshwater red alga Audouinella macrospora (Florideophyceae, Rhodophyta) and related forms based on SSU rRNA gene sequence analysis and pit plug ultrastructure. Journal of Phycology, 36(2): 433-440.
DOI:10.1046/j.1529-8817.2000.99173.x |
Ronquist F, Teslenko M, Van Der Mark P, Ayres D L, Darling A, Höhna S, Larget B, Liu L, Suchard M A, Huelsenbeck J P. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61(3): 539-542.
DOI:10.1093/sysbio/sys029 |
Saunders G W, Bird C J, Ragan M A, Rice E L. 1995. Phylogenetic relationships of species of uncertain taxonomic position within the Acrochaetiales-Palmariales complex (Rhodophyta): inferences from phenotypic and 18s rDNA sequence data. Journal of Phycology, 31(4): 601-611.
DOI:10.1111/j.1529-8817.1995.tb02556.x |
Saunders G W, Jackson C, Salomaki E D. 2018. Phylogenetic analyses of transcriptome data resolve familial assignments for genera of the red-algal Acrochaetiales-Palmariales Complex (Nemaliophycidae). Molecular Phylogenetics and Evolution, 119: 151-159.
DOI:10.1016/j.ympev.2017.11.002 |
Saunders G W. 1993. Gel purification of red algal genomic DNA: an inexpensive and rapid method for the isolation of polymerase chain reaction-friendly DNA. Journal of Phycology, 29(2): 251-254.
DOI:10.1111/j.0022-3646.1993.00251.x |
Saunders G W. 2005. Applying DNA barcoding to red macroalgae: a preliminary appraisal holds promise for future applications. Philosophical transactions of the Royal Society B: Biological Sciences, 360(1462): 1879-1888.
DOI:10.1098/rstb.2005.1719 |
Scott F J, Saunders G W, Kraft G T. 2013. Entwisleia bella, gen. et sp. nov., a novel marine "batrachospermaceous" red alga from southeastern Tasmania representing a new family and order in the Nemaliophycidae. European Journal of Phycology, 48(4): 398-410.
DOI:10.1080/09670262.2013.849359 |
Sheath R G, Müller K M. 1999. Systematic status and phylogenetic relationships of the freshwater genus Balbiania (Rhodophyta). Journal of Phycology, 35(4): 855-864.
DOI:10.1046/j.1529-8817.1999.3540855.x |
Sheath R G, Vis M L. 2015. Red algae. In: Wehr J D, Sheath R G, Kociolek J eds. Freshwater algae of North America. 2nd ed. Academic Press, San Diego, California. p. 237-264, https://doi.org/10.1016/B978-0-12-385876-4.00005-0.
|
Shi Z X. 2006. Flora algarum sinicarum aquae dulcis, Tomus XIII, Rhodophyta, Phaeophyta. Science Press, Beijing. 208p.
(in Chinese)
|
Skinner S, Entwisle T J. 2001. Non-marine algae of Australia: 3. Audouinella and Balbiania (Rhodophyta). Telopea, 9(3): 713-723.
DOI:10.7751/telopea20024009 |
Skuja H. 1944. Untersuchungen über die Rhodophyceen des Süsswassers (VII-XII). Acta Horti Botanici Universitatis Latviensis, 14: 3-64.
|
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28(10): 2731-2739.
DOI:10.1093/molbev/msr121 |
Thiers B. 2021. (Constantly updated). Index Herbariorum: A Global Directory of Public Herbaria and Associated Staff. New York Botanical Garden's Virtual Herbarium, New York. https://sweetgum.nybg.org/ih/. Accessed on Apr. 25, 2021.
|
Vis M L, Saunders G W, Sheath R G, Dunse K, Entwisle T J. 1998. Phylogeny of the Batrachospermales (Rhodophyta) inferred from rbcL and 18S ribosomal DNA gene sequences. Journal of Phycology, 34(2): 341-350.
DOI:10.1046/j.1529-8817.1998.340341.x |
Vis M L, Sheath R G. 1997. Biogeography of Batrachospermum gelatinosum (Batrachospermales, Rhodophyta) in North America based on molecular and morphological data. Journal of Phycology, 33(3): 520-526.
DOI:10.1111/j.0022-3646.1997.00520.x |
Volovsek M E, Hommersand M H, Manos P. 2000. The phylogenetics of Acrochaetiales revisited: inferences from rbcL and chloroplast encoded 23s rRNA sequence analysis. Journal of Phycology, 36(S3): 68-69.
DOI:10.1046/j.1529-8817.1999.00001-203.x |
Xie S L, Shi Z X. 2004. Balbiania Sirodot — a new record of freshwater red algae from China. Acta Botanica Boreali-Occidentalia Sinica, 24(9): 1732-1733.
(in Chinese with English abstract) DOI:10.3321/j.issn:1000-4025.2004.09.030 |
Zhang D, Gao F L, Jakovlić I, Zou H, Zhang J, Li W X, Wang G T. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Molecular Ecology Resources, 20(1): 348-355.
DOI:10.1111/1755-0998.13096 |
 2022, Vol. 40
2022, Vol. 40


