Institute of Oceanology, Chinese Academy of Sciences
Article Information
- PAN Chen, LIAO Zhi, HE Jianyu, GU Zhongqi, WANG Chunyue, FAN Meihua, BUTTINO Isabella, GUO Baoying, YAN Xiaojun, ZHANG Xiaolin
- Carnosine concentration and expression profiles of carnosine related genes in Mytilus after beta-alanine injection
- Journal of Oceanology and Limnology, 40(3): 1121-1134
- http://dx.doi.org/10.1007/s00343-021-1078-x
Article History
- Received Mar. 22, 2021
- accepted in principle Apr. 16, 2021
- accepted for publication May. 21, 2021
2 Bureau of Science and Technology Shengsi, Zhoushan 316022, China;
3 Italian Institute for Environmental Protection and Research (ISPRA), Via Vitaliano Brancati 48, Rome 00144, Italy
As a dipeptide with important biological functions and potential therapeutic uses, carnosine (beta-alanyl-L-histidine) has been discovered in the early 1900s (Gulewitsch and Amiradžibi, 1900). Carnosine and related histidine-containing dipeptides (HCDs) play diverse functions throughout all kingdoms of life.Although several physiological properties of carnosine are relevant to muscular function and homeostasis (Boldyrev et al., 2013; Blancquaert et al., 2015), the biological role and metabolism of HCDs are unclear till now. In the past 100 years, the distribution of HCDs in different animal species has been the focus of research. As reviewed by Boldyrev et al. (2013), HCDs are probably only present in vertebrates, where carnosine (or anserine) is abundant in skeletal muscle, neuronal tissues and, to a smaller extent, in the heart, liver, and kidney (Mannion et al., 1992; Boldyrev et al., 2013). Nobody had ever detected HCDs in invertebrates or other eukaryotes, such as plants and fungi. On the other hand, the genes of the enzymes for carnosine synthesis (such as carnosine synthase, CARNS) had been detected in genomic data of several mollusks species, such as CARNS-like proteins from Crassostrea virginica(XP_022325739.1) and Mizuhopecten yessoensis (XP_021350145.1), indicating carnosine may also present in invertebrates, at least in mollusks.
The carnosine metabolism pathways are mainly involved in synthesis from and hydrolysis to its constituent amino acids, by CARNS and carnosinase (CNDP) respectively (Teufel et al., 2003; Drozak et al., 2010). The synthesis of carnosine is dependent on the availability of the beta-alanine in vivo (Hill et al., 2007), and beta-alanine has been shown to be the rate-limiting precursor for carnosine synthesis in human muscle cells (Harris et al., 2006). Accordingly, beta-alanine supplementation was found to increase muscle carnosine content significantly (Harris et al., 2006; Hill et al., 2007). Therefore, beta-alanine is fast becoming a popular ergogenic aid to sports performance in recent years (Harris et al., 2006; Hill et al., 2007). According to limited literatures, the supplementation of beta-alanine tended to increase body weight gain and feed intake of aquatic organisms, such as carp (Geda et al., 2015), highlighting a potential application of beta-alanine supplementation on aquaculture.
Mytilus coruscus is an edible mussel with important economic and nutritional value in the East China Sea. In previous works, transcriptomic analysis was performed for the mantle and the foot of M. coruscus, and several genes associated with carnosine metabolic pathways were predicted, such as CARNS, CNDP, beta-alanine transport (TauT), and beta-alanine transaminase (ABAT). Accordingly, we speculated that M. coruscus contains carnosine in its body, similar to that of vertebrates, and the carnosine may have specific biochemical and physiological functions in Mytilus. Therefore, the concentration of HCDs in M. coruscus tissues was determined using an amino acid analyzer. The results revealed the existence of HCD in all tested tissues of M. coruscus. In addition, representative genes for carnosine metabolism were analyzed by real-time quantitative PCR (qRT-PCR) to verify the expression of these genes in HCD metabolism in mussel. To explore the transcriptional events in mussel in response to beta-alanine stimuli, transcriptomic sequencing before and after beta-alanine injection was further performed and the expressional profiles of carnosine-related genes of M. coruscus were presented here. The study aims to elucidate for the first time the concentration and localization of HCDs, as well as to understand the possible metabolic pathway of these dipeptides in Mytilus.
2 MATERIAL AND METHOD 2.1 Quantification of HCD in M. coruscus tissuesAdults of M. coruscus (6–8-cm length) were collected from the East China Sea near to Zhejiang Province. The shells were removed and the tissues, including adductor muscle, foot, mantle, gonad, and gill, were collected respectively. The tissues were weighed and then smashed immediately using liquid nitrogen, and then homogenized by a handheld homogenizer in pure water. Intracellular HCDs were released from the homogenate by an ultrasonic cell crusher. Samples were deproteinized through 10% trichloroacetic acid and then centrifuged at 20 000×g for 5 min at 4 ℃. The supernatants were collected and ultra-filtrated by a 3-kD cutoff, and then transferred to vials and lyophilized for use. Free amino acids were determined with an amino acid analyzer (LA8080, Hitachi, Japan).
For amino acid analysis, an aluminium ion exchange column (4.6 mm×60.0 mm, Hitachi, Japan) was used for sample separation. The reaction column was a heat-conducting membrane stainless steel column (4.6 mm×(42.5+42.5) mm, Hitachi, Japan). Analyzing temperature was set at 50 ℃ for exchange column and 135 ℃ for reaction column, respectively. Flow rate was 0.4 mL/min for elution buffer and 0.35 mL/min for ninhydrin, respectively. The detection wavelength was set at 570 nm, and the detection time was 150 min. The standard amino acid mixture (AN-Ⅱ, Hitachi, Japan) was used for qualitative and quantitative analysis.
2.2 qRT-PCR analysis of genes related to carnosine metabolismAccording to the literature (Miyaji et al., 2012; Everaert et al., 2013), five genes were supposed to be involved in the metabolic process of carnosine, including CARNS, CNDP, carnosine/histidine transporters (PHT), TauT, and ABAT. The mRNA sequence of these five genes was screened from the transcriptomic data of M. coruscus and the specific primers (Supplementary Table S1) were designed after gene sequence verification for qRT-PCR analysis.
For the tissue-specific expression of the five genes, various tissues (mantle, adductor muscle, gonad, foot, and gill) were collected from the adults of M. coruscus. For the quantification of mRNA levels induced by beta-alanine injection at different time points, adults of M. coruscus were injected preliminarily into adductor muscle with 25 μL of beta-alanine solution with a concentration of 1 mg/mL dissolved in sterilized seawater. Five tissues were then collected at 0, 1, 3, 6, and 12 h post-injection, respectively. Total RNA was then extracted from the five tissue samples by TRIzol reagent, and the cDNA first strand was synthesized by the PrimeScriptTM RT kit (TaKaRa). Three independent replicates are performed in the qRT-PCR analyses by using SYBR® Premix Ex TaqTM (TaKaRa) on an MX3000P Real-Time PCR system (Stratagene, US). The reactions were initially denatured at 95 ℃ for 180 s, followed by 35 cycles of 95℃ for 20 s, 59 ℃ for 20 s, and 72 ℃ for 25 s. Specific primers derived from the five genes were used for qRT-PCR. A melting curve analysis was performed to assess the amplification specificity. The relative expression levels were measured using the 2-ΔΔCt method (Livak and Schmittgen, 2001) with β-actin as an internal reference. The statistical analysis of differences was performed with one-way ANOVA followed by post hoc Student-Newman-Keuls (SNK) test using GAD package in R (v4.0.2) environment. Differences were considered significant at P < 0.05.
2.3 Transcriptomic analysis of M. coruscus before and after beta-alanine injectionAfter being placed in a thermostatic water tank without feeding for 48 h, the adults of M. coruscus (6–8-cm length) were randomly divided into two groups (n=9 animals per group) and injected into the adductor muscle with 25 μL of beta-alanine solution with the same concentration for qRT-PCR analysis. At 6 h post-injection, the whole soft body of M. coruscus was collected and the total RNA was then extracted for sequencing.
Total RNA was extracted from the whole soft body using TRIzol® reagent following manual (Invitrogen, Carlsbard, CA, USA). Genomic DNA was removed by using DNase I (TaKaRa). The total RNA integrity and purity was determined by 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara CA, USA) and quantified using the ND-2000 (NanoDrop Thermo Scientific, Wilmington, DE, USA).
RNA purification, reverse transcription, library construction and sequencing were performed at Shanghai Majorbio Bio-pharm Biotechnology Co., Ltd. (Shanghai, China), in which, the RNA sequencing was performed in a Novaseq 6000 system according to the manufacturer's instructions (Illumina, San Diego, CA). Adaptor sequences in raw data were trimmed and reads with low quality or length less than 70 bp were further removed by SolexaQA software. After the removal of ambiguous nucleotides, duplicates, and low-quality sequences (Phred quality scores b20), cleaned reads were obtained. Clean reads were assembled using the Trinity software (v2012_10_05) with the default parameters (Grabherr et al., 2011). All the assembled transcripts were searched against the NCBI protein nonredundant (NR), Clusters of Orthologous Groups (COG), and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases using BLASTX to identify the proteins that had the highest sequence similarity with the given transcripts to retrieve their function annotations and a typical cut-off E-values less than 1.0×10-5 was set. The highest homology of the aligned results was used to determine the sequence direction of unigenes.
The high-quality clean reads of each RNA-seq library were mapped to the assembled transcripts with Bowtie program (Langmead, 2010). The expression level of each transcript was calculated according to the transcripts per million reads (TPM) method, and the differential expression genes (DEGs) between different samples were then identified through their TPM. Gene abundances were quantified by RSEM (http://deweylab.biostat.wisc.edu/rsem/) (Li and Dewey, 2011). Essentially, differential expression analysis was performed using the DESeq2 (Love et al., 2014) with Q value≤0.05, DEGs with|log2FC| > 1 and Q value≤0.05 were considered to be significantly different expressed genes. Gene ontology (GO) and KEGG functional-enrichment analysis were performed to identify which DEGs were significantly enriched in GO terms and metabolic pathways compared with the whole-transcriptome background, with the Bonferroni-corrected P-value≤0.05. GO and KEGG pathway functional enrichment analysis were carried out by Goatools (https://github.com/tanghaibao/Goatools) and KOBAS (http://kobas.cbi.pku.edu.cn/home.do) (Xie et al., 2011).
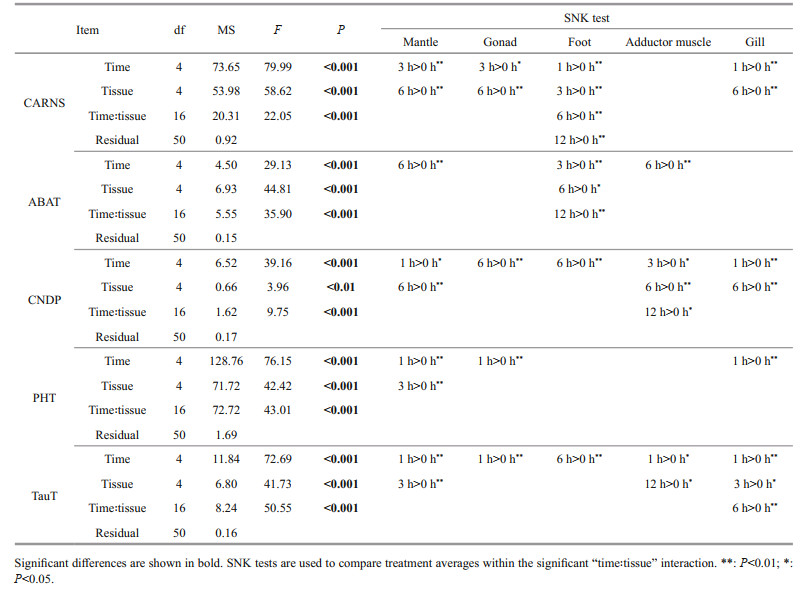
3 RESULT 3.1 Quantification of HCD in M. coruscus tissuesUsing an amino acid analyzer, the contents of free amino acids and HCDs (carnosine and anserine) in five tissues of M. coruscus were determined, and the contents of beta-alanine, histidine, carnosine, and anserine are shown in Table 1.
|
The results show that all the tested tissues contain HCDs and the content of anserine was significantly higher than that of carnosine (P < 0.05), indicating that the anserine was the dominant HCD in M. coruscus tissues (Table 1). Carnosine was detected only in the foot and gill with a low concentration (< 2-ng/g dry tissue). The adductor muscle contains the highest content of anserine (571.35-ng/g dry tissue), followed by the foot (252.79-ng/g dry tissue) and the mantle (47.01-ng/g dry tissue) (Table 1). We noticed that the content of anserine is relevant to the content of histidine and beta-alanine in various tissues. For example, the adductor muscle containing the highest content of anserine also contains the highest content of histidine and beta-alanine (Table 1).
At 6 h post-injection of beta-alanine, the content of anserine increased in most tested tissues except for the foot, in which, the content of anserine decreased from 252.79 ng/g to 134.01 ng/g (Table 1). In addition, for the mantle and the adductor muscle with no carnosine detected before beta-alanine injection, the content of carnosine can be detected with low concentration after beta-alanine injection (Table 1). These results indicated that the beta-alanine injection significantly increased the content of HCDs in most tissues of M. coruscus (P < 0.05).
3.2 mRNA expression profiles of carnosine-related enzymes and transporters in different Mytilus tissuesThe tissue-specific expression of five carnosine metabolic-related genes was investigated using qRT-PCR, and the five mRNAs were ubiquitously expressed in all tissues investigated, including the mantle, adductor muscle, gill, gonad, and foot (Fig. 1a). Both CARNS and CNDP was expressed with high level in the mantle, adductor muscle, and foot. The TauT (a beta-alanine transporters), was mainly expressed in muscular tissues (mantle, adductor muscle, and foot). Interestingly, we observed a positive correlation between TauT mRNA expression level and the content beta-alanine in adductor muscle, as displayed in Table 1 and Fig. 1a. Concerning carnosine transporters, the expression level of PHT (carnosine/histidine transporter) is lower than that of other genes in all tested tissues, as well as the ABAT, which is able to convert beta-alanine to malonate semi-aldehyde (deamination reaction).

|
| Fig.1 Expression of five carnosine metabolic-related genes in different Mytilus tissues a. expression profile of CARNS, TauT, PHT, ABAT, and CNDP in Mytilus gill, gonad, foot, adductor mussle, and mantle tissues. The color from white to red of each sub-box means the expression level from low to high. "**" indicates the gene has significant expression in the tissue. CARNS and TauT have the highest expression in mantle and adductor muscle respectively; b. the temporal expression pattern of five carnosine metabolic-related genes after beta-alanine injection. CARNS and CNDP reached the highest expression level in most tissues at 6 h post-injection. PHT and TauT reached the highest expression level in muscular tissues at 6 h post-injection, and 1 or 3 h for un-muscular tissues. ABAT has the highest expression level at 6 h post-injection for the adductor muscle and gill, and 3 h post-injection for the gonad and foot, and 1 h for the mantle. Quantification of transcripts was determinate by qRT-PCR with 2-ΔΔCt method. ANOVAs analysis of these data are listed in Table 2. |
The variation in expression level of carnosine metabolic-related genes after beta-alanine injection was further measured in five tissues. As shown in Fig. 1b and Table 2, the expression of CARNS and CNDP reached the highest level in most tissues at 6 h post-injection. While for carnosine metabolic-related transporters, the expression of PHT and TauT reached the highest level in muscular tissues (such as adductor muscle and foot) at 6 h post-injection, and 1 or 3 h for un-muscular tissues (such as gill and gonad). For ABAT, the highest level was presented at 6 h for the adductor muscle and gill, and 3 h for the gonad and foot, and 1 h for the mantle (Fig. 1b & Table 2).
We observed that the expression of genes in the key regulatory steps of carnosine metabolism, including beta-alanine uptake (TauT), carnosine synthesis (CARNS), and carnosine hydrolysis (CNDP), presented the highest level in most tissues at 6 h post-injection of beta-alanine (Fig. 1b & Table 2). Therefore, the time point of 6 h was selected for transcriptomic analysis of M. coruscus after beta-alanine injection.
3.3 Transcriptomic sequencing and annotationRNA-Seq was performed on the whole soft body samples from two groups, of which three beta-alanine injected groups (n=3, designated as bAGs) and three sterilized seawater injected groups (n=3, designated as CGs) of M. coruscus. In total, 39.2-Gb clean data were obtained, with an average of at least 6.38-Gb clean reads for each group, take up 97.9% of the raw reads. The alignment statistics results of the two groups showed about 71.96% mapped reads compared to reference genes (Supplementary Table S2). The raw transcriptomic sequences in the present study were deposited in the NCBI SRA database with an accession number of BioProject ID PRJNA648780.
The assembled unigenes were compared with NR, Swiss-prot, Pfam, COG, GO, and KEGG databases with an E-value cut-off of 10e-5. A total of 32 638 unigenes, accounting for 40.42% of the total number of unigenes (80 757), were annotated by at least one database, including 30 043 (37.2%) in NR, 17 897(22.16%) in Swiss-prot, 23 419 (29%) in Pfam, 19 443 (24.08%) in COG, 22 123 (27.39%) in GO, and 14 129 (17.5%) in KEGG (Supplementary Table S3). As shown in the E-value distribution profile of the top hits in the NR database, 72.5% of the mapped sequences showed significant homology (< 1.0e-20), and 73.58% of the sequences had similarities greater than 80% (Supplementary Fig.S1).
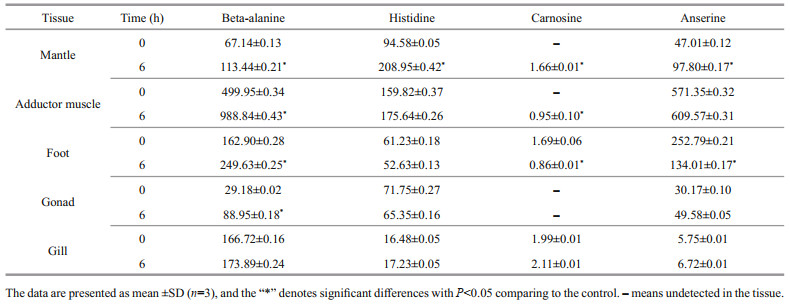
3.4 GO, COG, and KEGG functional classificationAs shown in Supplementary Fig.S2, the assignments to the cellular components were the most abundant (25 768 genes; 36.29%), followed by the molecular function (23 741 genes; 33.43%) and the biological process (21 503 genes; 30.28%). Cellular process (7 217 genes) was the most abundant category under the biological process class, followed by metabolic process, biological regulation, and cellular component organization or biogenesis. Under the cellular component class, the first three categories, membrane part, cell part, and organelle, accounted for approximately 69.42% of this classification. Under the molecular function class, binding (11 332 genes; 47.73%) and catalytic activity (8 303 genes; 34.97%) were the most categories. For COG annotation, most unigene (11 358; 54.8%) were annotated in the category of "function unknown", followed by "posttranslational modification, protein turnover, chaperones" (1 951; 9.4%), "intracellular trafficking, secretion, and vesicular transport" (1 529; 7.3%), and "signal transduction mechanisms" (766; 3.7%)(Supplementary Fig.S3). For KEGG annotation, 14 129 (17.5%) unigenes were significantly matched and assigned into 6 KEGG categories containing 43 pathways at second category level (Supplementary Fig.S4).
3.5 Differential expression genes (DEGs) analysisBased on the Benjamin-Hochberg method corrected P-adjust and the False Discovery Rate (FDR) analysis, the gene expression levels in bAGs were compared to that of CGs using DESeq2 software, and 3 569 DEGs (with fold change ≥2 and P-adjust < 0.05) were identified, including 2 245 up-regulated genes and 1 324 down-regulated genes (Fig. 2).

|
| Fig.2 The DEGs volcano plot The x-axis indicates the logarithm of gene expression fold-change, and the y-axis indicates the logarithm of P-adjust. The green and red dots denote downregulated and upregulated gene numbers, respectively, and the grey dots indicate the gene numbers with no significantly expression (Nosig) (with fold change < 2 and P-adjust > 0.05). |
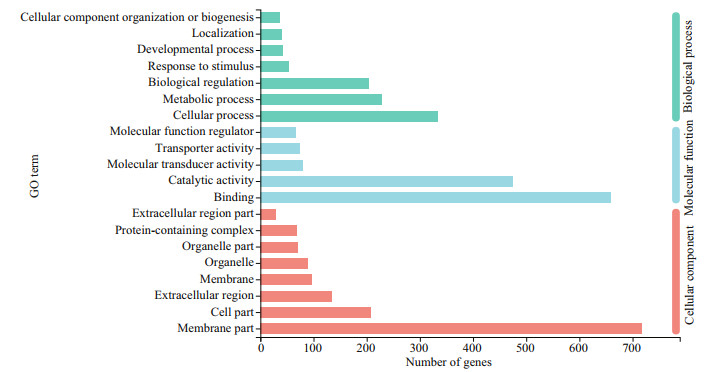
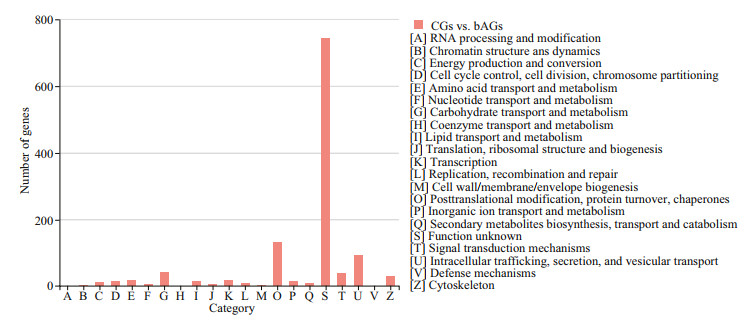
Functional annotation analysis of DEGs was performed in GO, COG, and KEGG databases, respectively. For GO annotation, 938, 1 356, and 1 415 DEGs were identified under biological process, molecular function, and cellular component, respectively (Fig. 3). The "cellular process" (333; 9.3%), "binding" (660; 18.5%), and "membrane part" (717; 20.1%) were subcategories with the most abundant genes in biological process, molecular function and cellular component, respectively.For COG classification, most DEGs (743; 60.3%) were assigned to the category of "Function unknown", followed by the category of "Posttranslational modification, protein turnover, and chaperones" (133; 10.1%), and the category of "Intracellular trafficking, secretion, and vesicular transport" (93; 7.5%) (Fig. 4). For KEGG analysis, 14 129 (17.5%) DEGs were annotated in the KEGG database, including 6 KEGG categories and 41 second categories, in which, the categories of "human diseases", "organismal systems", and "metabolism" were the top three pathways of DEGs (Fig. 5).
|
| Fig.3 The DEGs gene ontology (GO) classification Three main categories were summarized: biological process, molecular function, and cellular component. For the three categories, the top 7 functional groups under biological process and top 5 molecular function, and the 8 functional groups included in cellular component are presented, respectively. |
|
| Fig.4 DEGs COG annotation classification statistics The column subscripted with capital letters (A–Z) represents COG functional classification. The column height reflects the number of transcripts. |
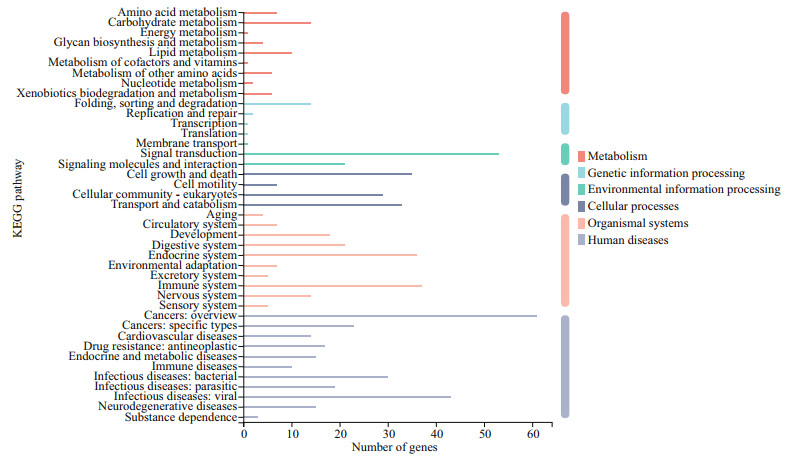
|
| Fig.5 KEGG annotation of DEGs Genes fell into six branches according to the KEGG metabolic pathways annotation: metabolism, genetic information processing, environmental information processing, cellular processes, organismal systems, and human diseases. |
GO and KEGG enrichment analysis were further performed using Goatools and Fisher validation. For GO enrichment, 1 406 DEGs were enriched, and the highest abundant DEGs presented in "cellular component", "membrane part", and "intrinsic component of membrane" of the cellular component category. For molecular function category, most DEGs were enriched in "molecular function regulator", "enzyme regulator activity", and "carbohydrate binding". In the biological process category, the DEGs were mainly enriched in "carbohydrate derivative metabolic process", "aminoglycan metabolic process", and "amino sugar metabolic process" (Supplementary Table S4). The top 20 GO enriched results of DEGs were shown in Fig. 6a.

|
| Fig.6 The top 20 enriched GO (a) functions and KEGG pathways (b) of DEGs FDR: false discovery rate. The x-axis indicate the rich factor of each GO function and KEGG pathway. The red dot represents the numbers of significant enriched DEGs. |
For KEGG enrichment analysis, a total of 770 DEGs were enriched in 24 KEGG pathways, in which, "focal adhesion", "human papillomavirus infection", and "proteoglycans in cancer" were the top three pathways with the highest abundant DEGs (Supplementary Table S5). The top 20 enriched KEGG pathways of DEGs were shown in Fig. 6b.
3.6 RNA-Seq confirmation by qRT-PCR analysisThe RNA-Seq data were validated by qRT-PCR analysis, and 10 randomly selected DEGs and three DEGs involving in carnosine-related metabolism were tested using specific primers (Supplementary Table S6). These genes included seven up-regulated and six down-regulated genes. The qRT-PCR results were highly coincident with the sequencing results (Fig. 7). Among them, CARNS (DN114), PHT(DN24997), and TauT (DN42305) genes were up-regulated, with an increased expression by 1–10 fold. These genes have similar expression patterns in RNA-Seq, and they can be considered as candidate beta-alanine induced genes. Similar gene expression trends were revealed in RNA-Seq data and qRT-PCR data.

|
| Fig.7 Validation of RNA-seq data by qRT-PCR Six significant down-regulated and seven up-regulated genes were selected to validate. The expression pattern of qRT-PCR were highly coincident with RNA-seq. |
Carnosine is an archetype of a family of HCDs that are found in the skeletal muscle and brain of vertebrate with some of the highest values reported in the wallaby, whale, marlin, and chicken (more than 10-mg/g tissue) (Boldyrev et al., 2013). Present study revealed that, M. coruscus also contains HCDs even at a low concentration (less than 1-μg/g tissue), and apparently the anserine is the primary HCD component in this mussel (Table 1). For fish, the relative higher concentration of HCDs in muscle was considered to be helpful for long-distance swimming of migratory pelagic fishes, such as marlin, trout, tuna, and salmon (Van Waarde, 1988). In present study, we also observed the relatively higher concentration of HCD in muscular tissues of M. coruscus, suggesting a similar function of HCD in muscle contraction. Although M. coruscus is a sessile species, we can not rule out that the presentation of HCD in this species may helpful for muscle contraction of the foot and the adductor muscle, considering that mussel foot is always active to sense something and to maintain byssus, and adductor muscle is used to open and shut shells. In addition, the higher concentration of anserine comparing to the carnosine in M. coruscus confirmed the observation that the anserine seems to be the predominant component of HCDs for most aquatic species (Boldyrev et al., 2013).
Beta-alanine is considered to be a performance-enhancing food supplement being used by competitive athletes to improve athletic performance (Blancquaert et al., 2015). Its efficacy has been summarized in a number of recent reviews (Trexler et al., 2015; Saunders et al., 2017), and recent research has reported the potential function of β-alanine supplementation in nervous system protection as well (Hoffman et al., 2019). Previous works highlighted the benefits of beta-alanine supplementation in human or other model animals (Ko et al., 2014), however, the effects of beta-alanine supplementation on invertebrates are still unknown. As a species with important economic value, M. coruscus culture has been on-going since 1980s in Zhoushan Islands, Zhejiang Province, China, and this industry has grown approximately 100-times, to more than 100 000 tons per year in 2019. Considering the benefits of beta-alanine supplementation on the livestock industry (Mei et al., 1998; Qi et al., 2018), we tested the expression profiles of carnosine metabolic-related genes in M. coruscus after beta-alanine injection.
From the literature, five possible regulatory steps were defined (Everaert et al., 2013), including beta-alanine uptake, carnosine synthesis, carnosine hydrolysis, carnosine and/or histidine transport, and other metabolic pathways of the beta-alanine. In this study, we identified mRNA expression of these five representative genes (CARNS, CNDP, TauT, PHT, and ABAT) by qRT-PCR in various tissues of M. coruscus. Our study demonstrates for the first time carnosine-related gene transcripts in different tissues of an invertebrate. To dissect the possible involvement of these genes, we further explored their expression in various interventions after beta-alanine injection. As summarized in Fig. 1, of the five carnosine-related genes, TauT, CARNS, CNDP were expressed with high level in muscular tissues rather than in gill and gonad of Mytilus, indicating that the carnosine is synthesized mainly in muscular tissues. Beta-alanine injection stimulated gene expression of TauT, CARNS, and CNDP, suggesting the abundant beta-alanine can be absorbed and transformed into carnosine in Mytilus tissues. These data were also in agreement with the results from the amino acids analyzer, in which, relative higher content of HCDs in muscular tissues was observed. The HCDs content was increased significantly after beta-alanine injection (Table 1). Similar results were also observed in other animals, such as mice (Everaert et al., 2013; Blancquaert et al., 2016). The expression of PHT (for carnosine transportation) showed a very low level in tested tissues. PHT has been reported as a transporter with carnosine and other oligopeptides, also L-histidine as its substrates (Daniel, 2004). The change in PHT mRNA expression after beta-alanine injection showed the highest sensitivity rather than other genes, particularly in the gonad, mantle, and gill. We thusly speculated that the PHT might be responsible for transportation of excess HCDs in the condition of beta-alanine injection. ABAT is an enzyme involved in oxidation (transamination) of beta-alanine (Blancquaert et al., 2016), and the low expression level of ABAT in Mytilus tissues and the slight up-regulation of this gene expression after beta-alanine injection suggested the priority role of beta-alanine is to serve as the precursor of carnosine synthesis. Yet, beta-alanine might be deaminated for further oxidation through ABAT in conditions of excess availability.
In this study, a high-throughput transcriptomic approach was implemented to explore the effects of beta-alanine injection on M. coruscus metabolism. According to the GO enrichment results of DEGs, we found 72 DEGs were enriched in the "carbohydrate derivative metabolic process" of biological process (Fig. 6a & Supplementary Table S4), indicating a strong relationship between beta-alanine and carbohydrate metabolism. As reported in previous works, beta-alanine supplementation can significantly increase the growth and feeding rate in fish (Ko et al., 2014; Geda et al., 2015), suggesting the roles of beta-alanine in macromolecules syntheses and degradation. Other studies have shown that the carnosine conversion efficiency of exogenous beta-alanine is only about 3%–6%, and the excess beta-alanine can be a carbon source for providing energy through oxidation (Pihl and Fritzson, 1955; Baxter and Roberts, 1961; Kurozumi et al., 1999; Ito et al., 2001; Rodionov et al., 2014). These findings indicated a potential application of beta-alanine in aquaculture, in which, the beta-alanine supplementation might be used for promoting the growth of aquatic animals.
A total of 770 DEGs were enriched in 24 KEGG pathways with most DEGs presented in the KEGG secondary categories of "cellular community", "infectious diseases", "cancers", "signaling molecules and interaction", "endocrine system", and "circulatory system" (Fig. 6b & Supplementary Table S5). These data indicated the complex effects of beta-alanine on M. coruscus metabolism, and those DEGs enriched in pathways of human diseases or cancers cannot be interpreted in present study. Interestingly, the ability of carnosine to inhibit tumor proliferation was confirmed in previous studies (Renner et al., 2010; Horii et al., 2012; Iovine et al., 2012), suggesting the possible roles of carnosine in cell proliferation. Therefore, we speculate that the elevated carnosine content in beta-alanine injected group may participate the cell proliferation-related pathways of Mytilus.
In addition, we found that the "vascular smooth muscle contraction" (map04270) was the most enriched pathway under the category of "circulatory system", as genes of this pathway were up-regulated after beta-alanine injection. It has been proven that carnosine plays a role in the contractile function of skeletal muscle in mammals (Culbertson et al., 2010; Bex et al., 2014), and potential roles of carnosine in muscles has been elucidated in previous works, including proton buffering capacity (Allen et al., 2008), a regulator of calcium release and calcium sensitivity (Zapata-Sudo et al., 1997; Dutka et al., 2012), and extracellular provider of histidine/histamine (Schröder et al., 2008). Our results suggested the potential roles of beta-alanine in the muscle function of Mytilus through carnosine. We also noticed that abundant DEGs were enriched in "endocrine system", including "oxytocin signaling pathway" (map04921), "melanogenesis" (map04916), and "renin secretion" (map04924), indicating that excess beta-alanine can activate some endocrine-related pathways of Mytilus to regulate organism homeostasis. There is some evidence to believe that carnosine can be synthesized and released from one tissue into other tissues for delivery of histidine or beta-alanine (Dunnett et al., 2002; Nagai et al., 2003). Thus, it has been suggested that carnosine could play an autocrine, paracrine, or even an endocrine role (Boldyrev et al., 2013).
Beta-alanine injection also changed the expression level of genes involving in amino acids metabolism in Mytilus, particularly in tyrosine metabolism. The relationship between beta-alanine and tyrosine has been elucidated in pervious study in invertebrates, such as Drosophila (Hodgetts and Choi, 1974), in which, beta-alanine plays a role in the tanning of adult fruit flies via preventing the extensive polymerization. However, although beta-alanine injection also showed effects on monoamine metabolism in rats, the levels of tyrosine and tryptophan did not show any significant change (Gomez et al., 1978). These results indicated different roles of beta-alanine in tyrosine metabolism between vertebrates and invertebrates. Beta-alanine injection significantly increased the expression level of genes involving in beta-alanine metabolism and histidine metabolism, particularly the CARNS, which is in agreement with the qRT-PCR data in present study. These results confirmed the observation from previous studies in vertebrates, in which, beta-alanine supplementation can elevate the expression level of carnosine related genes, as well as the content of HCDs (Bauer and Schulz, 1994; Harris et al., 2006; Derave et al., 2010).
Although our expression data are consistent with the existing literature and provide quantitative information on the tissue distribution of the carnosine relevant genes, the present study has some limitations. First, mRNA expression is not always consistent with the amount of mRNA that is effectively translated into protein. Therefore, our mRNA expression data should be considered as a preliminary study for the estimation of carnosine-related protein expression and function. Second, although the effects of beta-alanine injection on the expression of carnosine-related genes are demonstrated in the presented study, all the experiments were based on an acute supplementation of beta-alanine. Therefore, a long-term feeding of beta-alanine during mussel farming is necessary to evaluate the potential effect of beta-alanine on mussel physiology at a production scale.
5 CONCLUSIONIn summary, the results of present study suggest that M. coruscus contains HCDs with low concentration, and beta-alanine injection significantly induced the HCDs syntheses and the mRNA expression of carnosine-related genes. Transcriptomic profiling further revealed beta-alanine induced temporal expression patterns in mussel, highlighting the roles of beta-alanine on metabolism-related gene expression. This research provides insight into the beta-alanine-response mechanisms of mussel.
6 DATA AVAILABILITY STATEMENTThe transcriptomic data of this study has been submitted to NCBI with BioProject ID PRJNA648780; the cDNA sequences of M. coruscus carnosine synthase (CNARS), carnosinase (CNDP), beta-alanine transporter (TauT), and beta-alanine transaminase (ABAT), were submitted to GenBank with accession number QKX08451.1, QKX08453.1, QKX08456.1, QKX08455.1, and CAC5361819.1.
Electronic supplementary materialSupplementary material (Supplementary Tables S1–S6 and Figs.S1–S4) is available in the online version of this article at https://doi.org/10.1007/s00343-021-1078-x.
Allen D G, Lamb G D, Westerblad H. 2008. Skeletal muscle fatigue: cellular mechanisms. Physiological Reviews, 88(1): 287-332.
DOI:10.1152/physrev.00015.2007 |
Bauer K, Schulz M. 1994. Biosynthesis of carnosine and related peptides by skeletal muscle cells in primary culture. European Journal of Biochemistry, 219(1-2): 43-47.
DOI:10.1111/j.1432-1033.1994.tb19912.x |
Baxter C F, Roberts E. 1961. Elevation of γ-aminobutyric acid in brain: selective inhibition of γ-aminobutyric-α-ketoglutaric acid transaminase. Journal of Biological Chemistry, 236(12): 3287-3294.
DOI:10.1016/S0021-9258(18)94012-3 |
Bex T, Chung W, Baguet A, Stegen S, Stautemas J, Achten E, Derave W. 2014. Muscle carnosine loading by beta-alanine supplementation is more pronounced in trained vs. untrained muscles. Journal of Applied Physiology, 116(2): 204-209.
DOI:10.1152/japplphysiol.01033.2013 |
Blancquaert L, Baba S P, Kwiatkowski S, Stautemas J, Stegen S, Barbaresi S, Chung W, Boakye A A, Hoetker J D, Bhatnagar A, Delanghe J, Vanheel B, Veiga-Da-cunha M, Derave W, Everaert I. 2016. Carnosine and anserine homeostasis in skeletal muscle and heart is controlled by β-alanine transamination. Journal of Physiology, 594(17): 4849-4863.
DOI:10.1113/JP272050 |
Blancquaert L, Everaert I, Derave W. 2015. Beta-alanine supplementation, muscle carnosine and exercise performance. Current Opinion in Clinical Nutrition and Metabolic Care, 18(1): 63-70.
DOI:10.1097/MCO.0000000000000127 |
Boldyrev A A, Aldini G, Derave W. 2013. Physiology and pathophysiology of carnosine. Physiological Reviews, 93(4): 1803-1845.
DOI:10.1152/physrev.00039.2012 |
Culbertson J Y, Kreider R B, Greenwood M, Cooke M. 2010. Effects of beta-alanine on muscle carnosine and exercise performance: a review of the current literature. Nutrients, 2(1): 75-98.
DOI:10.3390/nu2010075 |
Daniel H. 2004. Molecular and integrative physiology of intestinal peptide transport. Annual Review of Physiology, 66(1): 361-384.
DOI:10.1146/annurev.physiol.66.032102.144149 |
Derave W, Everaert I, Beeckman S, Baguet A. 2010. Muscle carnosine metabolism and β-alanine supplementation in relation to exercise and training. Sports Medicine, 40(3): 247-263.
DOI:10.2165/11530310-000000000-00000 |
Drozak J, Veiga-da-Cunha M, Vertommen D, Stroobant V, Van Schaftingen E. 2010. Molecular identification of carnosine synthase as ATP-grasp domain-containing protein 1 (ATPGD1). Journal of Biological Chemistry, 285(13): 9346-9356.
DOI:10.1074/jbc.M109.095505 |
Dunnett M, Harris R C, Dunnett C E, Harris P A. 2002. Plasma carnosine concentration: diurnal variation and effects of age, exercise and muscle damage. Equine Veterinary Journal Supplement, 34(S34): 283-287.
DOI:10.1111/j.2042-3306.2002.tb05434.x |
Dutka T L, Lamboley C R, McKenna M J, Murphy R M, Lamb G D. 2012. Effects of carnosine on contractile apparatus Ca2+ sensitivity and sarcoplasmic reticulum Ca2+ release in human skeletal muscle fibers. Journal of Applied Physiology, 112(5): 728-736.
DOI:10.1152/japplphysiol.01331.2011 |
Everaert I, De Naeyer H, Taes Y, Derave W. 2013. Gene expression of carnosine-related enzymes and transporters in skeletal muscle. European Journal of Applied Physiology, 113(5): 1169-1179.
DOI:10.1007/s00421-012-2540-4 |
Geda F, Declercq A, Decostere A, Lauwaerts A, Wuyts B, Derave W, Janssens G P J. 2015. β-Alanine does not act through branched-chain amino acid catabolism in carp, a species with low muscular carnosine storage. Fish Physiology and Biochemistry, 41(1): 281-287.
DOI:10.1007/s10695-014-0024-7 |
Gomez M A M, Carlsson A, de Yebenes J G. 1978. The effect of β-alanine on motor behaviour, body temperature and cerebral monoamine metabolism in rat. Journal of Neural Transmission, 43(1): 1-9.
DOI:10.1007/BF02029014 |
Grabherr M G, Haas B J, Yassour M, Levin J Z, Thompson D A, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q D, Chen Z H, Mauceli E, Hacohen N, Gnirke A, Rhind N, Palma F D, Birren B W, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology, 29(7): 644-652.
DOI:10.1038/nbt.1883 |
Gulewitsch W, Amiradžibi S. 1900. Ueber das carnosin, eine neue organische base des fleischextractes. Berichte Der Deutschen Chemischen Gesellschaft, 33(2): 1902-1903.
DOI:10.1002/cber.19000330275 |
Harris R C, Tallon M J, Dunnett M, Boobis L, Coakley J, Kim H J, Fallowfield J L, Hill C A, Sale C, Wise J A. 2006. The absorption of orally supplied β-alanine and its effect on muscle carnosine synthesis in human vastus lateralis. Amino Acids, 30(3): 279-289.
DOI:10.1007/s00726-006-0299-9 |
Hill C, Harris R C, Kim H J, Harris B D, Sale C, Boobis L H, Kim C K, Wise J A. 2007. Influence of β-alanine supplementation on skeletal muscle carnosine concentrations and high intensity cycling capacity. Amino Acids, 32(2): 225-233.
DOI:10.1007/s00726-006-0364-4 |
Hodgetts R, Choi A. 1974. β alanine and cuticle maturation in Drosophila. Nature, 252(5485): 710-711.
DOI:10.1038/252710a0 |
Hoffman J R, Gepner Y, Cohen H. 2019. β-Alanine supplementation reduces anxiety and increases neurotrophin expression in both young and older rats. Nutrition Research, 62: 51-63.
DOI:10.1016/j.nutres.2018.11.001 |
Horii Y, Shen J, Fujisaki Y, Yoshida K, Nagai K. 2012. Effects of l-carnosine on splenic sympathetic nerve activity and tumor proliferation. Neuroscience Letters, 510(1): 1-5.
DOI:10.1016/j.neulet.2011.12.058 |
Iovine B, Iannella M L, Nocella F, Pricolo M R, Bevilacqua M A. 2012. Carnosine inhibits KRAS-mediated HCT116 proliferation by affecting ATP and ROS production. Cancer Letters, 315(2): 122-128.
DOI:10.1016/j.canlet.2011.07.021 |
Ito S, Ohyama T, Kontani Y, Matslida K, Sakata S F, Tamaki N. 2001. Influence of dietary protein levels on β-alanine aminotransferase expression and activity in rats. Journal of Nutritional Science and Vitaminology, 47(4): 275-282.
DOI:10.3177/jnsv.47.275 |
Ko R, Low Dog T, Gorecki D K, Cantilena L R, Costello R B, Evans W J, Hardy M L, Jordan S A, Maughan R J, Rankin J W, Smith-Ryan A E, Valerio L G, Jones D, Deuster P, Giancaspro G I, Sarma N D. 2014. Evidence-based evaluation of potential benefits and safety of beta-alanine supplementation for military personnel. Nutrition Reviews, 72(3): 217-225.
DOI:10.1111/nure.12087 |
Kurozumi Y, Abe T, Yao W B, Ubuka T. 1999. Experimental beta-alaninuria induced by (aminooxy) acetate. Acta Medica Okayama, 53(1): 13-18.
DOI:10.18926/AMO/31647 |
Langmead B. 2010. Aligning short sequencing reads with Bowtie. Current Protocols in Bioinformatics. Chapter 11: Unit 11.7. p. 1-24, https://doi.org/10.1002/0471250953.bi1107s32.
|
Li B, Dewey C N. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics, 12(1): 323.
DOI:10.1186/1471-2105-12-323 |
Livak K J, Schmittgen T D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods, 25(4): 402-408.
DOI:10.1006/meth.2001.1262 |
Love M I, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology, 15(12): 550.
DOI:10.1186/s13059-014-0550-8 |
Mannion A, Jakeman P, Dunnett M, Harris R C, Willan P L T. 1992. Carnosine and anserine concentrations in the quadriceps femoris muscle of healthy humans. European Journal of Applied Physiology and Occupational Physiology, 64(1): 47-50.
DOI:10.1007/BF00376439 |
Mei L, Cromwell G, Crum A, Decker E A. 1998. Influence of dietary β-alanine and histidine on the oxidative stability of pork. Meat Science, 49(1): 55-64.
DOI:10.1016/s0309-1740(97)00108-3 |
Miyaji T, Sato M, Maemura H, Takahata Y, Morimatsu F. 2012. Expression profiles of carnosine synthesis-related genes in mice after ingestion of carnosine or β-alanine. Journal of the International Society of Sports Nutrition, 9(1): 15.
DOI:10.1186/1550-2783-9-15 |
Nagai K, Niijima A, Yamano T, Otani H, Okumra N, Tsuruoka N, Nakai M, Kiso Y. 2003. Possible role of L-carnosine in the regulation of blood glucose through controlling autonomic nerves. Experimental Biology and Medicine, 228(10): 1138-1145.
DOI:10.1177/153537020322801007 |
Pihl A, Fritzson P. 1955. The catabolism of C14-labeled β-alanine in the intact rat. Journal of Biological Chemistry, 215(1): 345-351.
DOI:10.1016/S0021-9258(18)66042-9 |
Qi B, Wang J, Ma Y B, Wu S G, Qi G H, Zhang H J. 2018. Effect of dietary β-alanine supplementation on growth performance, meat quality, carnosine content, and gene expression of carnosine-related enzymes in broilers. Poultry Science, 97(4): 1220-1228.
DOI:10.3382/ps/pex410 |
Renner C, Zemitzsch N, Fuchs B, Geiger K D, Hermes M, Hengstler J, Gebhardt R, Meixensberger J, Gaunitz F. 2010. Carnosine retards tumor growth in vivo in an NIH3T3-HER2/neu mouse model. Molecular Cancer, 9(1): 2.
DOI:10.1186/1476-4598-9-2 |
Rodionov R N, Jarzebska N, Weiss N, Lentz S R. 2014. AGXT2: a promiscuous aminotransferase. Trends in Pharmacological Sciences, 35(11): 575-582.
DOI:10.1016/j.tips.2014.09.005 |
Saunders B, Elliott-Sale K, Artioli G G, Swinton P A, Dolan E, Roschel H, Sale C, Gualano B. 2017. β-alanine supplementation to improve exercise capacity and performance: a systematic review and meta-analysis. British Journal of Sports Medicine, 51(8): 658-669.
DOI:10.1136/bjsports-2016-096396 |
Schröder L, Schmitz C H, Bachert P. 2008. Carnosine as molecular probe for sensitive detection of Cu (Ⅱ) ions using localized 1H NMR spectroscopy. Journal of Inorganic Biochemistry, 102(2): 174-183.
DOI:10.1016/j.jinorgbio.2007.07.033 |
Teufel M, Saudek V, Ledig J P, Bernhardt A, Boularand S, Carreau A, Cairns N J, Carter C, Cowley D J, Duverger D, Ganzhorn A J, Guenet C, Heintzelmann B, Laucher V, Sauvage C, Smirnova T. 2003. Sequence identification and characterization of human carnosinase and a closely related non-specific dipeptidase. Journal of Biological Chemistry, 278(8): 6521-6531.
DOI:10.1074/jbc.M209764200 |
Trexler E T, Smith-Ryan A E, Stout J R, Hoffman J R, Wilborn C D, Sale C, Kreider R B, Jäger R, Earnest C P, Bannock L, Campbell B, Kalman D, Ziegenfuss T N, Antonio J. 2015. International society of sports nutrition position stand: Beta-Alanine. Journal of the International Society of Sports Nutrition, 12(1): 30.
DOI:10.1186/s12970-015-0090-y |
Van Waarde A. 1988. Biochemistry of non-protein nitrogenous compounds in fish including the use of amino acids for anaerobic energy production. Comparative Biochemistry and Physiology Part B: Comparative Biochemistry, 91(2): 207-228.
DOI:10.1016/0305-0491(88)90136-8 |
Xie C, Mao X Z, Huang J J, Ding Y, Wu J M, Dong S, Kong L, Gao G, Li C Y, Wei L P. 2011. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Research, 39(S2): 316-322.
DOI:10.1093/nar/gkr483 |
Zapata-Sudo G, Sudo R T, Lin M, Nelson T E. 1997. Calcium-sensitizing function for the dipeptide carnosine in skeletal muscle contractility. Cellular Physiology and Biochemistry, 7(2): 81-92.
DOI:10.1159/000154861 |
 2022, Vol. 40
2022, Vol. 40