Institute of Oceanology, Chinese Academy of Sciences
Article Information
- WANG Jingqian, GAO Xinming, ZHENG Xuebin, DU Chen, HOU Congcong, XIE Qingping, LOU Bao, LIU Feng, JIN Shan, ZHU Junquan
- Evidence for the role of KIF17 in fish spermatid reshaping: expression pattern of KIF17 in Larimichthys polyactis spermiogenesis
- Journal of Oceanology and Limnology, 39(6): 2322-2335
- http://dx.doi.org/10.1007/s00343-020-0304-2
Article History
- Received Aug. 23, 2020
- accepted in principle Oct. 27, 2020
- accepted for publication Nov. 26, 2020
2 Zhejiang Academy of Agricultural Sciences, Hangzhou 310021, China
Spermiogenesis, or the post-meiotic development of spermatogenic cells, is a highly complex but orderly process. During spermiogenesis, spermatid reshaping occurs, in which round spermatids form streamlined sperm by a series of changes, including acrosome and tail formation, nuclear reshaping, and the disposal of extra cytoplasm (Wong-Riley and Besharse, 2012; Ma et al., 2017). Disruptions in spermatid reshaping result in poor sperm quality and even male infertility (Yan, 2009). Therefore, the molecular mechanisms underlining spermatid reshaping are crucial for understanding spermiogenesis and a focus of reproductive biology research.
Kinesins are a class of microtubule-dependent motor proteins (Hirokawa et al., 2009) with important roles in spermatid reshaping. Kinesins are highly conserved during evolution and transport many different cargos along the microtubule to the acrosome, the sperm tail, and the midpiece during spermiogenesis (Olson and Winfrey, 1990; Lehti et al., 2013; Ma et al., 2017). They may interact with the manchette, a transient perinuclear microtubule structure involved in spermatid nuclear shaping and sperm tail formation (Kierszenbaum, 2001; Lehti and Sironen, 2016; Ma et al., 2017).
The homodimeric KIF17, and the heterotrimeric kinesin-Ⅱ complexes, namely KIF3A/3B/KAP3 and KIF3A/3C/KAP3, constitute the kinesin-2 family in vertebrates (Silverman and Leroux, 2009). Homodimeric KIF17 is formed by the dimerization of identical polypeptide chains, both containing a pair of N-terminal head motor domains, a stalk domain containing two coiled-coil regions (CC1-CC2), and a C-terminal tail domain (Grüter et al., 1998; Miki et al., 2005; MacAskill et al., 2009). KIF17 mainly functions in the transport of different cargos in an adenosine triphosphate (ATP)-dependent manner toward the plus ends of microtubules to the tips of dendrites, cilia, or outer segments of photoreceptor cells (Setou et al., 2000; Chu et al., 2006; Kayadjanian et al., 2007; Takano et al., 2007; Insinna et al., 2009). In mice and rats, KIF17 is highly expressed in male germ cells and is localized in the manchette and the portion of the sperm tail implicated in spermiogenesis (Setou et al., 2000; Kierszenbaum, 2002; Macho et al., 2002; Chennathukuzhi et al., 2003; Kierszenbaum and Tres, 2004; Kimmins et al., 2004). Therefore, KIF17 probably contributes to nuclear shaping and tail formation by interacting with the manchette in mammals. However, the function of KIF17 in fish spermiogenesis has not been reported.
Larimichthys polyactis, a benthopelagic fish species in the family Sciaenidae, is an important economic fishery resource (Zhang et al., 2016). Therefore, studies of spermiogenesis in L. polyactis are crucial to obtain a theoretical basis for its breeding and culture. Kang et al. (2013) and Wang et al. (2019) examined the ultrastructural characteristics of spermiogenesis in L. polyactis. In early spermatid, the nucleus is oval or round, and the mitochondria randomly distributed in the perinuclear cytoplasm. In middle spermatid, the nucleus is round, and the nuclear chromatin is condensed into clumps. The mitochondria fuse and migrate to one side of the nucleus, where the sperm midpiece forms. At this time, the flagellum extends from the implantation fossa. In late spermatid, the nucleus is kidney-shaped, the nuclear chromatin is more condensed, the residual cytoplasm migrates gradually to the bottom of the implantation fossa, and the flagellum extends gradually. In mature sperm, the nuclear chromatin is highly condensed, and mitochondria are typically found in the sperm midpiece. Overall, L. polyactis spermiogenesis involves nuclear reshaping, tail formation, and the removal of excess cytoplasm (Fig. 1). These cytological changes of the spermatids during L. polyactis spermiogenesis are similar to those in many fish species (Zhang et al., 2017; Zhao et al., 2017). However, it is not clear that the molecular mechanism underlying spermatid reshaping during spermiogenesis in L. polyactis and in other taxa.
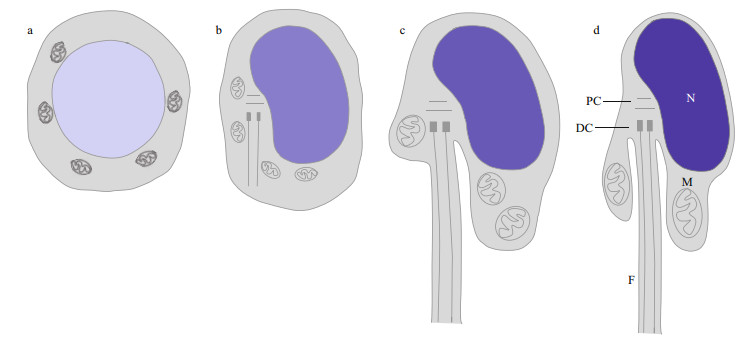
|
| Fig.1 Spermiogenesis in L. polyactis a. early spermatid; b. middle spermatid; c. late spermatid; d. mature sperm. N: nucleus; F: flagellum; M: mitochondria; PC: proximal centromere; DC: distal centromere. |
To investigate the functions of KIF17 in spermatid reshaping during L. polyactis spermiogenesis, we performed cDNA cloning and analyzed the tissue distribution and expression of Lp-kif17, as well as the colocalization of Lp-KIF17 and microtubules. Our results provide evidence that Lp-KIF17 is involved in spermatid reshaping and, in particular, may promote nuclear reshaping and tail formation during spermiogenesis.
2 MATERIAL AND METHOD 2.1 Tissue samplingThe male L. polyactis individuals were sampled from Zhoushan (Zhejiang, China). Seven tissues (the testis (Ⅱ–Ⅴ stage), intestine, heart, brain, muscle, liver, and kidney) were extracted and stored at -80 ℃. Experiments were approved by the Animal Ethics Committee of Ningbo University and strictly enforced according to the Experimental Animal Management Law of China.
2.2 RNA extraction and reverse transcriptionTotal tissue RNA was extracted from each tissue type of L. polyactis using the TRIzol Reagent (Invitrogen, San Diego, CA, USA). Reverse transcription for real-time quantitative PCR (qPCR) was performed using the PrimeScript® RT Reagent Kit (TaKaRa, Dalian, China). the synthesis of first-strand cDNA of 5′ Rapid Amplification of cDNA Ends (RACE) and 3′ RACE were performed using the Smart RACE cDNAAmplification Kit (Clontech, Mountain View, CA, USA) and 3′ Full RACE Amplification Kit (TaKaRa, Kusatsu, Japan), respectively. All cDNAs were stored at -20 ℃.
2.3 Cloning of full-length kif17 cDNAThe open reading frame (ORF) of Lp-kif17 was obtained from the full-length genome (ASM1011929v1). The specific primers (Table 1) for 5′ RACE and 3′ RACE were designed base on the ORF sequence of kif17 cDNA. The cDNA products were separated, and the target bands were extracted, purified, ligated, transformed and ultimately sequenced using GENEWIZ (Suzhou, China).
The Lp-KIF17 sequence was predicted using the Sequence Manipulation Suite (http://www.bio-soft.net/sms/index.html). Multiple alignments of Lp-KIF17 amino acid sequences were generated using Vector NTI10. A phylogenetic tree of KIF17 was constructed by the neighbor-joining method using MEGA 5.0. The functional domains and tertiary structure of KIF17 proteins were predicted using the ProtParam tool (http://web.expasy.org/protparam/) and I-TASSER (http://zhanglab.ccmb.med.umich.edu/I-TASSER/), respectively.
2.5 Quantitative analysis of Lp-kif17 mRNA expressionThe mRNA expression levels of kif17 in the heart, intestine, muscle, liver, brain, kidney, and testis (Ⅱ-Ⅴ) were measured by qPCR, as described previously (Zhao et al., 2018). Lp-kif17-specific primers (Table 1) were designed using Primer Premier 5 and synthesized by GENEWIZ (Suzhou, China). The β-actin (Actb) gene was chosen as a positive control. The relative mRNA levels of Lp-kif17 were analyzed using the 2-ΔΔCt method. Statistical analyses performed with SPSS 16.0 were using one-way analysis of variance (ANOVA) followed by the Duncan's multiple comparison, where P < 0.05 indicated a significant difference.
2.6 Fluorescence in situ hybridization (FISH)The Ⅳ stage testes of male L. polyactis were extracted, and frozen sections were produced and preserved as described previously (Zhao et al., 2017). The kif17 probe (Table 1) was synthesized by GENEWIZ (Suzhou, China) and was conjugated to fluorescein with fluorescein isothiocyanate (FITC); Probe specificity was evaluated using the BLAST. The experiment was conducted based on the methods of Zhao et al. (2018). The fluorescence signal were observed and photographed using a confocal laser-scanning microscope (LSM880; Carl Zeiss, Oberkochen, Germany).
2.7 AntibodiesA mouse anti-Lp-KIF17 antibody was prepared as reported by Gao et al. (2019). The sequence encoding the tail domain of KIF17 (aa 567–725) was amplified, and the polypeptide was approximately 17.6 kDa. The cDNA sequence was inserted into the pEASY-Blunt E2 vector (TransGen, Beijing, China) to construct the prokaryotic expression vector. The primers Anti-KIF17-F is GACACCAGGCAGAGGAAGAAC and the primers Anti-KIF17-R is TCTGTTGATGTTG-GGGATGCA. The constructed pEASY-Blunt E2-KIF17 plasmid was transformed into Transetta (DE3) competent cells. The transformed cells were incubated until the OD600 reached 0.6, and the expression of KIF17 was induced by isopropyl-β-d-thiogalactopyranoside (IPTG) at a final concentration of 1 mmol/L. The recombinant protein KIF17 was purified using the His-tag Protein Purification Kit (Beyotime, Shanghai, China) following the manufacturer's instructions. Finally, the KIF17 recombination protein was used for ICR mouse immunization to produce the antiserum according to Gao et al. (2019).
2.8 Western blottingThe quality of the mouse anti-Lp-KIF17 antibody was tested by western blot analysis, following a previously described method (Zhao et al., 2018). HRP-conjugated goat anti-mouse IgG (H+L) (Beyotime) was used as the secondary antibody. Protein bands were visualized by chemiluminescence imaging (Tanon 5200; Shanghai, China).
2.9 Immunofluorescence stainingThe IF experiment was conducted according to Wang et al. (2019). The frozen tissue sections were incubated with a mouse anti-KIF17 antibody (diluted 1:70) and rabbit anti-tubulin antibody (diluted 1:100; Beyotime) at 4 ℃ overnight. Then the sections were incubated with Alexa Fluor 555-labeled Goat Anti-Mouse IgG (H+L) (Beyotime) and Alexa Fluor 488-labeled Donkey Anti-Rabbit IgG (H+L) (Beyotime), for 1 h at 37 ℃. The immunostained tissues sections were mounted and observed using a confocal laser-scanning microscope. The control group was processed without the addition of the primary antibody.
3 RESULT 3.1 Characterization of the full-length cDNA of Lp-kif17 and the Lp-KIF17 proteinThe cloned kif17 cDNA had a 332-bp 5′ untranslated region (UTR), 480-bp 3′ UTR, and 2 433-bp ORF encoding 810 amino acids (aa) (Fig. 2). The predicted molecular weight of Lp-KIF17 protein was 90.3 kDa. The deduced Lp-KIF17 protein possessed three domains: a motor domain (3–343 aa), a stalk domain (344–652 aa) containing two coiled-coil regions (398–439 aa/615–653 aa), and a tail domain (654– 810 aa) (Fig. 3b). Furthermore, the tertiary structure of Lp-KIF17 was predicted (Fig. 3c), including a motor domain (Fig. 3f), a stalk domain containing two coiled-coil regions (Fig. 3g), and a tail domain (Fig. 3h). As in other kinesins, Lp-KIF17 had putative ATP hydrolysis sites (Fig. 3d) and microtubule-binding motifs (Fig. 3e) in the motor domain. A model of the putative Lp-KIF17 homodimeric complex is depicted in Fig. 3i.

|
| Fig.2 Full-length cDNA of Lp-kif17 and the deduced amino acid sequence Nucleotides are numbered in the 5′→3′ direction. The initiation codon was ATG, and the termination codon was TAA, which is marked in red. The deduced amino acid sequence is shown underneath the nucleotide sequence. The full-length cDNA of Lp-kif17 was 3 245 bp, consisting of a 332-bp 5′ UTR, a 2 433-bp ORF encoding 810 aa, and a 480-bp 3′ UTR. |

|
| Fig.3 Multiple alignments and structure prediction of Lp-KIF17 a. multiple alignments of KIF17 amino acid sequences of L. polyactis and other species. The same amino acids are represented in yellow regions; more than 50% similarity are represent in blue regions, and a lower similarity are represent in green regions. The motor domain is marked in a red box. Green triangles indicate microtubule-binding sites. Red triangles indicate putative ATP hydrolysis sites. The species names and the GenBank accession numbers of KIF17 were as follows: Mus musculus (NP_001177907.1); Notechis scutatus (XP_026532176.1); Xenopus laevis (XP_018097355.1); Danio rerio (XP_001919146.1); Carassius auratus (XP_026130843.1); Larimichthys crocea (XP_027135593.1); Pomacea canaliculata (XP_025086307.1); b. protein structure domain of Lp-KIF17; c–h. the predicted tertiary structure of Lp-KIF17; d. the positions of the predicted ATP hydrolysis sites; e. the positions of the predicted microtubule-binding sites; f. the predicted N-terminal motor domain (red); g. the predicted stalk domain (blue); h. the predicted C-terminal tail domain (orange); i. model pattern of the putative Lp-KIF17 homodimeric complex. Head: the N-terminal head motor domain; CC1: coiled-coil region 1; CC2: coiled-coil region 2; tail: the C-terminal tail domain. |
We aligned the Lp-KIF17 amino acid sequence with sequences of homologs in other species (Fig. 3). The sequence identities between the Lp-KIF17 sequence and sequences of homologs in Mus musculus, Notechis scutatus, Xenopus laevis, Danio rerio, Carassius auratus, Larimichthys crocea, and Pomacea canaliculata were 52.8%, 56.7%, 55.6%, 66.2%, 67.1%, 94.4%, and 74.7%, respectively. As shown in Fig. 4, a phylogenetic analysis showed that Lp-KIF17 formed a group with orthologues in other teleosts, distinct from other kinesin-2 family clusters. Moreover, Lp-KIF17 was most closely related to the KIF17 of L. crocea.
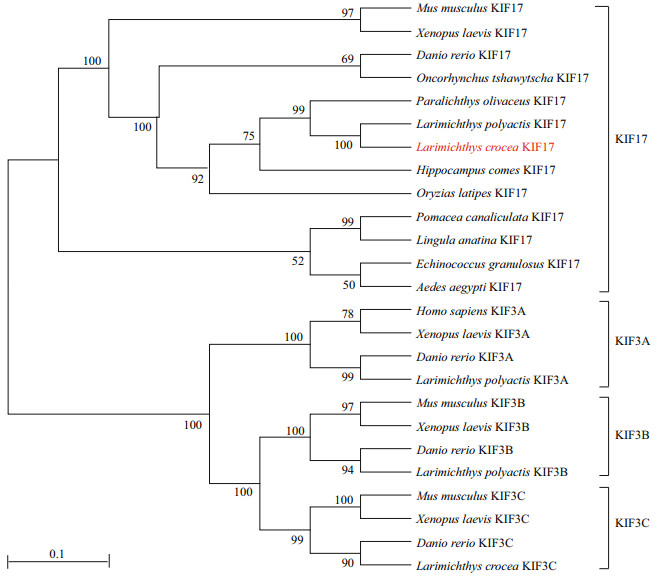
|
| Fig.4 Phylogenetic tree of KIF17 and 25 available kinesin-2 proteins The phylogenetic tree was generated based on the alignment of amino acid sequences using MEGA 5.0 software using the neighbor-joining method. The amino acid sequences were obtained from the GenBank and the accession numbers were as follows: Homo sapiens: KIF3A (NP_001287720.1); Danio rerio: KIF17 (XP_001919146.1), KIF3A (NP_001017604.2), KIF3B (NP_001093615.1), KIF3C (XP_002661420.3); Xenopus laevis: KIF17 (XP_018097355.1), KIF3A (XP_018111667.1), KIF3B (XP_018092567.1), KIF3C (XP_018121538.1); Larimichthys crocea KIF17 (XP_027135593.1), KIF3C (XP_010738232.2); Echinococcus granulosus KIF17 (XP_024349597.1), Paralichthys olivaceus KIF17 (XP_019952108.1); Hippocampus comes KIF17 (XP_019712241.1); Oryzias latipes KIF17 (XP_023811060.1); Oncorhynchus tshawytscha KIF17 (XP_024282861.1), Pomacea canaliculata KIF17 (XP_025086307.1), Lingula anatina KIF17 (XP_013396997.1), Aedes aegypti KIF17 (XP_021693306.1), Larimichthys polyactis: KIF3A (QBH87852.1), KIF3B (QBH87853.1); Mus musculus: KIF3B (NP_032470.3), KIF3C (NP_032471.2), KIF17 (NP_034753.1). |
As determined by qPCR, Lp-kif17 expression showed a wide distribution in all tested tissues, with high expression level in the brain and testis and low expression level in other tissue types (Fig. 5a). We further evaluated Lp-kif17 expression during Ⅱ–Ⅴ stages of testis development (Fig. 5b). The mRNA expression of Lp-kif17 increased gradually, peaking at stage Ⅳ, and then decreased dramatically at stage Ⅴ.
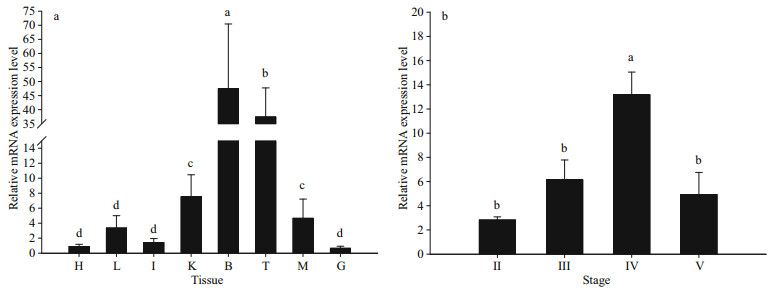
|
| Fig.5 Quantitative analysis of Lp-kif17 mRNA a. the relative expression level of kif17 mRNA to β-actin in the heart (H), liver (L), intestine (I), kidney (K), brain (B), testis (T), muscle (M), and gill (G) of L. polyactis; b. relative expression level of kif17 mRNA to β-actin in different stages of testis development. Values are represented as mean±S.D. Different lowercase letters mean significant differences (P < 0.05) among the different groups, respectively. |
The expression and distribution of Lp-kif17 were evaluated by FISH (Fig. 6). In early spermatid, the Lp-kif17 mRNA signals were evenly distributed in the perinuclear cytoplasm (Fig. 6a1–a4). During middle spermatid, the signals were enhanced and were concentrated around the nucleus (Fig. 6b1–b4). In late spermatid, the signals mainly accumulated on one side of the nucleus where the tail forms (Fig. 6c1–c4). In mature sperm, mRNA was detected at the midpiece of sperm (Fig. 6d1–d4).

|
| Fig.6 Fluorescence in-situ hybridization results of Lp-kif17 mRNA during spermiogenesis in L. polyactis Blue signals represent nuclei stained with 4′, 6-diamidino-2-phenylindole (DAPI). Lp-kif17 mRNA was stained with FITC labeled probe (green). a1, b1, c1, d1 show images of spermiogenesis in the hematoxylin-eosin (HE) stain; a2–a4: early spermatid; b2–b4: middle spermatid; c2–c4: late spermatid; d2–d4: sperm. The white virtual box indicates an enlarged image in the region of arrow. The scale bars are 5 μm. |
Lp-KIF17 was successfully expressed in Escherichia coli Transetta (DE3) (Fig. 7a). A clear target band was found in induced cells (line 3) but not in un-induced cells (line 2) by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The purified recombinant protein showed a single band (line 4) with an approximate molecular mass of 17.6 kDa. Finally, the recombination protein Lp-KIF17 was used for mouse immunization, and the antiserum was extracted as a mouse anti-KIF17 polyclonal antibody against L. polyactis. Western blotting was used to evaluate the specificity of this antibody. A ~90-kDa protein band was only detected (Fig. 7b), consistent with the predicted molecular weight of Lp-KIF17. These results suggest that the mouse anti-KIF17 polyclonal antibody could be used for subsequent IF assays.
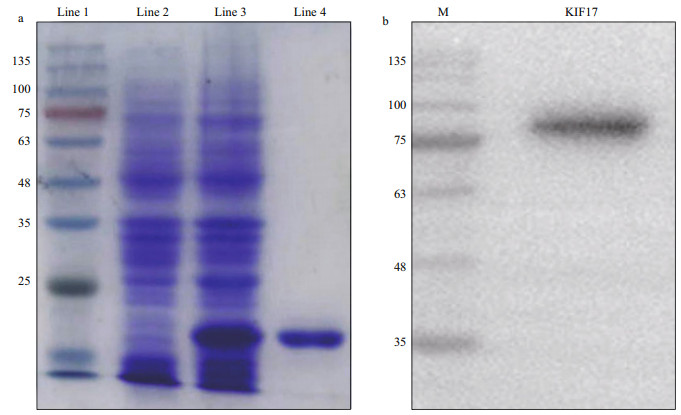
|
| Fig.7 Expression and purification of recombinant protein Lp-KIF17 and specificity assessment of mouse anti-Lp-KIF17 antibodie a. the expression and purification of recombinant protein Lp-KIF17. Line 1: molecular markers; line 2: un-induced total protein; line 3: total protein with KIF17 expression induced by IPTG; line 4: purified recombination protein with a molecular weight of approximately 17.6 kDa; b. mouse anti-Lp-KIF17 antibody assessment by western blotting. M: protein markers. |
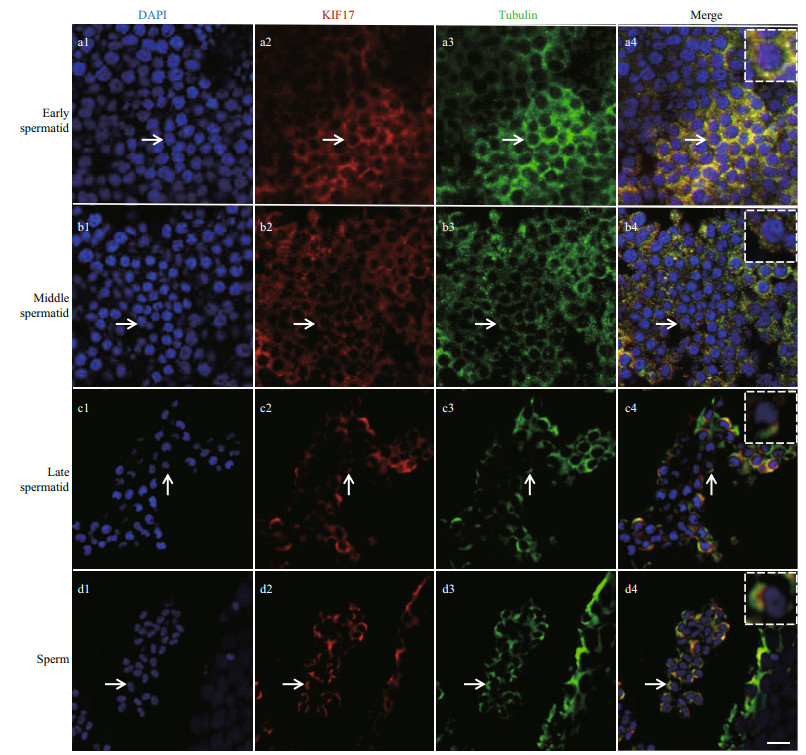
As determined by immunofluorescence, Lp-KIF17 was expressed and colocalized with microtubules during spermiogenesis in L. polyactis. In early spermatid, Lp-KIF17 and tubulin were evenly distributed in the cytoplasm (Fig. 8a1–a4). In middle spermatid, the localization patterns of Lp-KIF17 and tubulin signals were similar to those of early spermatid and accumulated around the nucleus (Fig. 8b1–b4). In late spermatid, the signals migrated to one side of the nucleus where sperm tail forms (Fig. 8c1–c4). In mature sperm, Lp-KIF17 and tubulin were mainly detected in the midpiece of the sperm (Fig. 8d1–d4). No signals were detected in negative control group (data not shown).
|
| Fig.8 Immunofluorescence localization of Lp-KIF17 and tubulin during spermiogenesis in L. polyactis Nuclei were stained with DAPI (blue). Microtubules were stained with Alexa Fluor 488-labeled antibody (green). Lp-KIF17 was stained with an Alexa Fluor 555-labeled antibody (red). a1–a4. early spermatid; b1–b4. middle spermatid; c1–c4. late spermatid; d1–d4. sperm. The white virtual box indicates an enlarged image in the region of arrow. The scale bar is 5 μm. |
KIF17 is a homodimeric motor protein belonging to the kinesin-2 family (Silverman and Leroux, 2009). The KIF17 domain structure is well-characterized in mammals (Yamazaki et al., 1995; Miki et al., 2005; MacAskill et al., 2009). The motor domain functions in movement along microtubules by binding to microtubules and hydrolyzing ATP. The tail domain is a highly variable domain that binds to various cargos. The stalk region, containing two coiled-coil domains (CC1-CC2), is a regulatory region controller (Milic et al., 2017). The structural characteristics of KIF17 were similar to those of other kinesin-2 family members (KIF3A, KIF3B, and KIF3C) and N-terminal kinesins. However, the stalk domains of KIF3A, KIF3B, KIF3C, and other kinesins usually have a single coiled-coil domain, whereas KIF17 has two coiled-coil domains. The special structural features of the stalk domain of KIF17 may be related to its auto-inhibition by folding around its coiled-coil domain to suppress motility in the absence of cargo (Imanishi et al., 2006; Hammond et al., 2010). In this study, a full-length cDNA sequence was obtained for kif17 from the testis of L. polyactis. The predicted Lp-KIF17 protein possessed all of the main structural features of previously-reported KIF17, including an N-terminal head motor domains containing microtubule-binding and ATP hydrolysis sites, a stalk domain containing two coiled-coil regions (CC1–CC2), and a C-terminal tail domain. Multiple sequence alignment of KIF17 proteins in various taxa illustrated that Lp-KIF17 is relatively structurally conserved and that its motor domain is more highly conserved than its non-motor domains. The high conservation of the motor domain may reflect its involvement in ATP hydrolysis and microtubule-binding; the C-terminal domain, on the contrary, is involved in interaction with various cargos (Miki et al., 2005). The structural conservatism of Lp-KIF17 suggests that its function was conserved during evolution.
In L. polyactis, kif17 mRNA levels were highest in the brain, followed by the testis, and were low in other tissues. This expression pattern is consistent with previous results (Nakagawa et al., 1997; Setou et al., 2000). KIF17 in the brain is likely to function in the transport of neuronal proteins to dendrites in a microtubule-dependent manner. In the mouse, high KIF17 expression in the central nervous system may be related to the transport of N-methyl-d-aspartate receptor NR2B subunit, potassium Kv4.2 channels, and kainate receptor GluR5 from cell bodies to dendrites (Setou et al., 2000; Chu et al., 2006; Kayadjanian et al., 2007). In Caenorhabditis elegans, Osm-3 (a homolog of mammalian KIF17) shows highly specific expression in neurons and is thought to contribute to dendritic transport (Tabish et al., 1995). Hence, Lp-kif17 in the brain may be related to dendritic transport. In addition, high KIF17 expression in the testis may be related to spermiogenesis (Wong-Riley and Besharse, 2012; Ma et al., 2017). In mice, KIF17 is highly expressed in the testis and may participate in spermiogenesis by interacting with the manchette to promote nuclear reshaping and tail formation (Saade et al., 2007). Similarly, the high expression of kif17 in the testis suggests that KIF17 is involved in L. polyactis spermiogenesis. In this study, we further determined that kif17 expression in the testis is highest at stage Ⅳ. In this stage, the testis of L. polyactis contains abundant spermatids, which undergo morphological transformation into spermatozoa by spermiogenesis (Kang et al., 2013). Therefore, we speculate that Lp-kif17 in the testis may be related to spermiogenesis. To confirm our conjecture, we tracked the spatiotemporal expression pattern of Lp-kif17 mRNA by FISH during spermiogenesis. The Lp-kif17 mRNA signals were continuously detected throughout spermiogenesis. Overall, these results suggested that kif17 is involved in L. polyactis spermiogenesis.
During spermatid reshaping, the nucleus undergoes striking chromosome condensation and compression, eventually leading to nuclear elongation and tail formation (Ma et al., 2017). During spermiogenesis, a transient perinuclear microtubule structure, called the manchette, appears when the round nucleus begins to elongate and gradually moves toward the posterior half of the spermatid to elongate the nucleus by the formation of a connection between the microtubule and the nuclear skeleton. This structure disappears when the elongation and condensation of the spermatid nucleus are near completion (Kierszenbaum, 2002). Previous studies have shown that the manchette plays a significant role in spermatid nuclear shaping and tail formation by sorting structural and functional molecules during nuclear shaping and sperm tail development (Kierszenbaum, 2002; Kierszenbaum and Tres, 2004). In azh (abnormal sperm head shape) mouse mutants, the abnormal position of the manchette is correlated with an abnormally-shaped spermatid nucleus and multiple coiling of the sperm tail (Mochida et al., 1999). The manchette displays complex motility via the activity of several kinesins. For instance, KIF3A knockout mice show abnormally manchettes and typical malformed heads, suggesting that kinesin KIF3A participates in spermatid reshaping by interacting with the manchette (Lehti et al., 2013). In mice, KIF17 protein signals are colocalized with the manchette, further supporting the relationship between KIF17 and the manchette (Saade et al., 2007). In our immunofluorescence analysis of L. polyactis, tubulin signals were evenly distributed in the perinuclear cytoplasm in the early spermatid stage, primarily distributed around the nucleus in the middle spermatid stage, and distributed on the side where the midpiece forms in the late spermatid stage. In mature spermatozoa, the signals of tubulin were mainly detected in the midpiece of the sperm. The localization pattern of tubulin suggests that L. polyactis may have a perinuclear microtubule structure, similar to the manchette, involved in spermatid nuclear shaping. We further found that Lp-KIF17 colocalized with tubulin during early spermatid development into mature sperm, and the signals shifted from the perinuclear cytoplasm to one side where the midpiece forms, suggesting that Lp-KIF17 participates in spermatid nuclear shaping and tail formation during spermatid reshaping by interacting with perinuclear microtubules. Based on these findings, models of Lp-KIF17 involved during spermiogenesis is prosed (Fig. 9).

|
| Fig.9 Overview potential function of Lp-KIF17 during spermiogenesis KIF17 may participate in the spermatid nuclear shaping and tail formation by interacting with perinuclear microtubules during L. polyactis spermiogenesis. Head: the N-terminal head motor domain; CC1: coiled-coil region 1; CC2: coiled-coil region 2; tail: the C-terminal tail domain. |
In this study, we cloned the Lp-kif17 cDNA and characterized the expression patterns and potential functions of KIF17 during L. polyactis spermiogenesis. Lp-kif17 was widely expressed in most tissues, with the highest level in brain, followed by that in the testis. Furthermore, kif17 was continuously detected during spermiogenesis, suggesting that it plays a key role in this process. Lp-KIF17 colocalized with tubulin at all stages, and the signals moved from the perinuclear cytoplasm to the side of tail formation. These findings suggest that KIF17 may be involved in L. polyactis spermiogenesis and may specifically participate in nuclear shaping and tail formation by interacting with perinuclear microtubules during spermatid reshaping. Based on these findings, models of Lp-KIF17 involved during spermiogenesis is prosed. However, detailed function of KIF17 in L. polyactis spermiogenesis requires further study.
6 DATA AVAILABILITY STATEMENTThe data that support the findings of this study are available from the corresponding author upon reasonable request.
Chennathukuzhi V, Morales C R, El-Alfy M, Hecht N B. 2003. The kinesin KIF17b and RNA-binding protein TB-RBP transport specific cAMP-responsive element modulator-regulated mRNAs in male germ cells. Proceedings of the National Academy of Sciences of the United States of America, 100(26): 15566-15571.
DOI:10.1073/pnas.2536695100 |
Chu P J, Rivera J F, Arnold D B. 2006. A role for KIF17 in transport of Kv4.2. Journal of Biological Chemistry, 281(1): 365-373.
DOI:10.1074/jbc.M508897200 |
Gao X M, Mu D L, Hou C C, Zhu J Q, Jin S, Wang C L. 2019. Expression and putative functions of KIFC1 for nuclear reshaping and midpiece formation during spermiogenesis of Phascolosoma esculenta. Gene, 683: 169-183.
DOI:10.1016/j.gene.2018.10.021 |
Grüter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, Bachi A, Wilm M, Felber B K, Izaurralde E. 1998. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Molecular Cell, 1(5): 649-659.
DOI:10.1016/S1097-2765(00)80065-9 |
Hammond J W, Blasius T L, Soppina V, Cai D W, Verhey K J. 2010. Autoinhibition of the kinesin-2 motor KIF17 via dual intramolecular mechanisms. The Journal of Cell Biology, 189(6): 1013-1025.
DOI:10.1083/jcb.201001057 |
Hirokawa N, Noda Y, Tanaka Y, Niwa S. 2009. Kinesin superfamily motor proteins and intracellular transport. Nature Reviews Molecular Cell Biology, 10(10): 682-696.
DOI:10.1038/nrm2774 |
Imanishi M, Endres N F, Gennerich A, Vale R D. 2006. Autoinhibition regulates the motility of the C. elegans intraflagellar transport motor OSM-3. The Journal of Cell Biology, 174(7): 931-937.
DOI:10.1083/jcb.200605179 |
Insinna C, Humby M, Sedmak T, Wolfrum U, Besharse J C. 2009. Different roles for KIF17 and kinesin Ⅱ in photoreceptor development and maintenance. Developmental Dynamics, 238(9): 2211-2222.
DOI:10.1002/dvdy.21956 |
Kang H W, Chung E Y, Chung J S, Lee K Y. 2013. Ultrastructural studies of spermatogenesis and the functions of Leydig cells and Sertoli cells associated with spermatogenesis in Larimichthys polyactis (Teleostei, Perciformes, Sciaenidae). Animal Cells and Systems, 17(4): 250-258.
DOI:10.1080/19768354.2013.829783 |
Kayadjanian N, Lee H S, Pina-Crespo J, Heinemann S F. 2007. Localization of glutamate receptors to distal dendrites depends on subunit composition and the kinesin motor protein KIF17. Molecular and Cellular Neuroscience, 34(2): 219-230.
DOI:10.1016/j.mcn.2006.11.001 |
Kierszenbaum A L, Tres L L. 2004. The acrosome-acroplaxome-manchette complex and the shaping of the spermatid head. Archives of Histology and Cytology, 67(4): 271-284.
DOI:10.1679/aohc.67.271 |
Kierszenbaum A L. 2001. Spermatid manchette: plugging proteins to zero into the sperm tail. Molecular Reproduction and Development, 59(4): 347-349.
DOI:10.1002/mrd.1040 |
Kierszenbaum A L. 2002. Intramanchette transport (IMT): managing the making of the spermatid head, centrosome, and tail. Molecular Reproduction and Development, 63(1): 1-4.
DOI:10.1002/mrd.10179 |
Kimmins S, Kotaja N, Fienga G, Kolthur U S, Brancorsini S, Hogeveen K, Monaco L, Sassone-Corsi P. 2004. A specific programme of gene transcription in male germ cells. Reproductive Biomedicine Online, 8(5): 496-500.
DOI:10.1016/s1472-6483(10)61094-2 |
Lehti M S, Kotaja N, Sironen A. 2013. KIF3A is essential for sperm tail formation and manchette function. Molecular and Cellular Endocrinology, 377(1-2): 44-55.
DOI:10.1016/j.mce.2013.06.030 |
Lehti M S, Sironen A. 2016. Formation and function of the manchette and flagellum during spermatogenesis. Reproduction, 151(4): R43-R54.
DOI:10.1530/REP-15-0310 |
Ma D D, Wang D H, Yang W X. 2017. Kinesins in spermatogenesis. Biology of Reproduction, 96(2): 267-276.
DOI:10.1095/biolreprod.116.144113 |
MacAskill A F, Rinholm J E, Twelvetrees A E, Arancibia-Carcamo I L, Muir J, Fransson A, Aspenstrom P, Attwell D, Kittler J T. 2009. Miro1 is a calcium sensor for glutamate receptor-dependent localization of mitochondria at synapses. Neuron, 61(4): 541-555.
DOI:10.1016/j.neuron.2009.01.030 |
Macho B, Brancorsini S, Fimia G M, Setou M, Hirokawa N, Sassone-Corsi P. 2002. CREM-Dependent transcription in male germ cells controlled by a kinesin. Science, 298(5602): 2388-2390.
DOI:10.1126/science.1077265 |
Miki H, Okada Y, Hirokawa N. 2005. Analysis of the kinesin superfamily: insights into structure and function. Trends in Cell Biology, 15(9): 467-476.
DOI:10.1016/j.tcb.2005.07.006 |
Milic B, Andreasson J O L, Hogan D W, Block S M. 2017. Intraflagellar transport velocity is governed by the number of active KIF17 and KIF3AB motors and their motility properties under load. Proceedings of the National Academy of Sciences of the United States of America, 114(33): E6830-E6838.
DOI:10.1073/pnas.1708157114 |
Mochida K, Tres L L, Kierszenbaum A L. 1999. Structural and biochemical features of fractionated spermatid manchettes and sperm axonemes of the Azh/Azh mutant mouse. Molecular Reproduction and Development, 52(4): 434-444.
DOI:10.1002/(SICI)1098-2795(199904)52:4<434::AID-MRD13>3.0.CO;2-D |
Nakagawa T, Tanaka Y, Matsuoka E, Kondo S, Okada Y, Noda Y, Kanai Y, Hirokawa N. 1997. Identification and classification of 16 new kinesin superfamily (KIF) proteins in mouse genome. Proceedings of the National Academy of Sciences of the United States of America, 94(18): 9654-9659.
DOI:10.1073/pnas.94.18.9654 |
Olson G E, Winfrey V P. 1990. Mitochondria-cytoskeleton interactions in the sperm midpiece. Journal of Structural Biology, 103(1): 13-22.
DOI:10.1016/1047-8477(90)90081-M |
Saade M, Irla M, Govin J, Victorero G, Samson M, Nguyen C. 2007. Dynamic distribution of Spatial during mouse spermatogenesis and its interaction with the kinesin KIF17b. Experimental Cell Research, 313(3): 614-626.
DOI:10.1016/j.yexcr.2006.11.011 |
Setou M, Nakagawa T, Seog D H, Hirokawa N. 2000. Kinesin superfamily motor protein KIF17 and mLin-10 in NMDA receptor-containing vesicle transport. Science, 288(5472): 1796-1802.
DOI:10.1126/science.288.5472.1796 |
Silverman M A, Leroux M R. 2009. Intraflagellar transport and the generation of dynamic, structurally and functionally diverse cilia. Trends in Cell Biology, 19(7): 306-316.
DOI:10.1016/j.tcb.2009.04.002 |
Tabish M, Siddiqui Z K, Nishikawa K, Siddiqui S S. 1995. Exclusive expression of C. elegans osm-3 kinesin gene in chemosnsory neurons open to the external environment. Journal of Molecular Biology, 247(3): 377-389.
DOI:10.1006/jmbi.1994.0146 |
Takano K, Miki T, Katahira J, Yoneda Y. 2007. NXF2 is involved in cytoplasmic mRNA dynamics through interactions with motor proteins. Nucleic Acids Research, 35(8): 2513-2521.
DOI:10.1093/nar/gkm125 |
Wang J Q, Gao X M, Zheng X B, Hou C C, Xie Q P, Lou B, Zhu J Q. 2019. Expression and potential functions of KIF3A/3B to promote nuclear reshaping and tail formation during Larimichthys polyactis spermiogenesis. Development Genes and Evolution, 229(4): 161-181.
DOI:10.1007/s00427-019-00637-5 |
Wong-Riley M T T, Besharse J C. 2012. The kinesin superfamily protein KIF17: one protein with many functions. Biomolecular Concepts, 3(3): 267-282.
DOI:10.1515/bmc-2011-0064 |
Yamazaki H, Nakata T, Okada Y, Hirokawa N. 1995. KIF3A/B: a heterodimeric kinesin superfamily protein that works as a microtubule plus end-directed motor for membrane organelle transport. The Journal of Cell Biology, 130(6): 1387-1399.
DOI:10.1083/jcb.130.6.1387 |
Yan W. 2009. Male infertility caused by spermiogenic defects: lessons from gene knockouts. Molecular and Cellular Endocrinology, 306(1-2): 24-32.
DOI:10.1016/j.mce.2009.03.003 |
Zhang B D, Xue D X, Wang J, Li Y L, Liu B J, Liu J X. 2016. Development and preliminary evaluation of a genomewide single nucleotide polymorphisms resource generated by RAD-seq for the small yellow croaker (Larimichthys polyactis). Molecular Ecology Resources, 16(3): 755-768.
DOI:10.1111/1755-0998.12476 |
Zhang D D, Gao X M, Zhao Y Q, Hou C C, Zhu J Q. 2017. The C-terminal kinesin motor KIFC1 may participate in nuclear reshaping and flagellum formation during spermiogenesis of Larimichthys crocea. Fish Physiology and Biochemistry, 43(5): 1351-1371.
DOI:10.1007/s10695-017-0377-9 |
Zhao Y Q, Mu D L, Wang D, Han Y L, Hou C C, Zhu J Q. 2018. Analysis of the function of KIF3A and KIF3B in the spermatogenesis in Boleophthalmus pectinirostris. Fish Physiology and Biochemistry, 44(3): 769-788.
DOI:10.1007/s10695-017-0461-1 |
Zhao Y Q, Yang H Y, Zhang D D, Han Y L, Hou C C, Zhu J Q. 2017. Dynamic transcription and expression patterns of KIF3A and KIF3B genes during spermiogenesis in the shrimp, Palaemon carincauda. Animal Reproduction Science, 184: 59-77.
DOI:10.1016/j.anireprosci.2017.06.017 |
 2021, Vol. 39
2021, Vol. 39



