Institute of Oceanology, Chinese Academy of Sciences
Article Information
- WANG Kai, WANG Xin, ZOU Qiang, JIANG Han, ZHANG Rongrong, TIAN Yanan, ZHANG Lele, LIN Qiang
- Genome-wide evolution of MAPKs family and their expression in response to bacterial infection in seahorse Hippocampus erectus
- Journal of Oceanology and Limnology, 39(6): 2309-2321
- http://dx.doi.org/10.1007/s00343-020-0332-y
Article History
- Received Aug. 25, 2020
- accepted in principle Nov. 9, 2020
- accepted for publication Nov. 25, 2020
2 CAS Key Laboratory of Tropical Marine Bio-resources and Ecology, South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou 510301, China;
3 Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou), Guangzhou 511458, China;
4 Yantai Branch of Shandong Technology Transfer Center, Chinese Academy of Sciences, Yantai 264003, China;
5 University of Chinese Academy of Sciences, Beijing 100049, China
The mitogen-activated protein kinases (MAPKs) are an evolutionarily conserved family of serine/threonine protein kinases, involved in the regulation of a variety of cellular processes including proliferation, differentiation, and apoptosis (Cargnello and Roux, 2011; Plotnikov et al., 2011; Arthur and Ley, 2013). Previous literatures have demonstrated that all eukaryotic cells possess multiple MAPK pathways to recognize and respond to extracellular stimuli, which correspondingly regulate various cellular activities including metabolism, motility, proliferation, and differentiation (Krens et al., 2006). The three major MAPK subfamilies, extracellular signal regulated kinase (erk), c-Jun NH2-terminal kinase (jnk), and the p38, were characterized as typical MAPKs in multicellular organisms (Sopontammarak et al., 2005). The MAPKs can phosphorylate their downstream substrates as transcription factors when they are activated by phosphorylation upon threonine and tyrosine residues with in a conserved Thr-Xaa-Tyr motif (Raman et al., 2007).
Environmental stresses and extracellular stimuli including hormones, growth factors, cytokine, transforming growth factor (TGF)-related agents that through Ser-Thr kinase receptors can activate MAPK signaling pathways (Kyriakis and Avruch, 2012). Once activated, the signal will transduced via the "core signaling module" consisting of a three-tier phosphorylation cascade driven sequentially by MAP kinase kinase kinases (MKKKs), MAP kinase kinases (MKKs) and MAP kinases (MAPKs) (Kyriakis and Avruch, 2012; Sun et al., 2016). At present, 14 mammalian MAPKs have been characterized into seven groups. Among these groups, the extracellular signal-regulated kinase 1 and 2 (erk1/2), the c-Jun N-terminal kinases 1-3 (jnk1-3), the p38s (p38α, β, γ, and δ), and erk5 are called the conventional MAPKs, while the other MAPKs (Nemo-like kinase (nlk), erk3/4, and erk7/8) sharing distinct mode of activation are classified as atypical MAPKs (Coulombe and Meloche, 2007; Cargnello and Roux, 2011; Plotnikov et al., 2011). Unlike the intensive concern in mammalian species, information about evolutionary origins, patterns and functions of the conventional MAPKs in teleost are limited.
Majority attention has been drawn to revealing how MAPKs functions in innate immunity, stress responses, development, and cellular functions in teleost species. It has been testified that JNKs participate in pattern recognition receptors (PRRs) induced inflammation and transduction of osmosensory signals (Kültz and Avila, 2001; Marshall et al., 2005; Ding et al., 2018), as well as promote cell regeneration (He et al., 2016). ERKs have fundamental roles in modulating cellular growth, mitosis, metabolism, and individual development (Codina et al., 2008; Krens et al., 2008; Fuentes et al., 2011; Sánchez-Gurmaches et al., 2013), and they have been found responding to PRRs induced immune challenges in Epiniphelus coioides, Larimichthys crocea, and Salmo salar (Iliev et al., 2013; Jia et al., 2015; Ding et al., 2018), as well as virus infection in S. salar (Olavarria et al., 2015). Moreover, ERKs can also respond to stresses, such as osmotic pressure, environmental neurotoxic metal, and thermal variation (Kültz and Avila, 2001; Leal et al., 2006; Keller et al., 2008; Jia et al., 2015). In aspect of immunologic functions, p38s can be activated by Lipopolysaccharide (LPS) and cytosine-phosphate-guanosine (CpG) (Iliev et al., 2013; Zhu et al., 2014; Olavarria et al., 2015). Activation induced by toxins, thermal, starvation, and osmotic pressure in many teleost fishes confirmed a functional role of p38 pathway in stress responses (Marshall et al., 2005; Leal et al., 2006; Urushibara et al., 2009; Feidantsis et al., 2012; Antonopoulou et al., 2013; Li et al., 2016; Marshall et al., 2017). Additionally, p38 involves in synchronous embryonic cleavage and myocardial regeneration in Danio rerio (Jopling et al., 2012). The members of MAPKs vary a lot in different teleost species, as well as the functional performance of an identical member. Therefore, our understanding of how MAPKs evolve and function in teleost is very necessary.
Seahorse is characterized by sedentary behavior with low swimming capacities, small home ranges, which evolves a unique morphology includes an armored body, a highly derived head shape, a prehensile tail, and the absence of caudal and pelvic fins (Lin et al., 2016; Luo et al., 2016). Its specialized life-history traits, including male-pregnancy with a brood pouch, low fecundity and high site fidelity, have attracted considerable attention in adaptive evolution and marine conservation (Foster and Vincent, 2004; Qin et al., 2017). The unique body morphology and specialized life history traits has made them good flagship species for threats and solutions in marine conservation (Zhang et al., 2017; Wang et al., 2019). In addition, the lack of spleen and gut-associated lymphatic tissue (GALT) may has resulted in a partial but natural "immunodeficiency" of seahorse (Matsunaga and Rahman, 1998; Galtier et al., 2009; Luo et al., 2016). Considering the vital functions of the MAPK gene family in immune response, we hypothesized that the adaptive evolution and expression characteristics of the MAPK gene family in the seahorse may have different characteristics from other teleost fishes. The draft genomes of the lined seahorse Hippocampus erectus provide a valuable information for molecular-evolutionary inference (Lin et al., 2017). It is possible for us to conduct systematic genomic analysis of MAPKs, as well as its correlations to the other known teleosts. The present study aimed to determine the conventional member, gene structure, evolutionary history, and expression pattern of MAPKs in the lined seahorse H. erectus. The results can enrich our knowledge of the evolution history of MAPKs, and will facilitate better understanding of the functional role of MAPKs in teleosts.
2 MATERIAL AND METHOD 2.1 Experimental animals and sample collectionHippocampus erectus were cultured in the aquaculture system of seahorse center in Ludong University, and all breeding methods were carried out in accordance with relevant guidelines and regulations of the Animal Care and Use Committee of the Ludong University. Lined seahorses were cultured in re-circulating seawater tanks (50 cm×40 cm×30 cm) with aerated water (salinity: 31.5±0.5, temperature: 25.5±0.5 ℃, and pH: 8.2±0.1). The seahorses were fed with frozen Mysis spp. three times a day (08:00, 12:00, and 16:00), as characterized by Luo et al. (2016). Thirty-six adult seahorses (body height: 12.7±0.8 cm, wet bodyweight: 13.1±0.8 g) were used in the Vibrio fortis challenge experiment. These seahorses were divided into three groups (12 inds./group) collected, and each group was reared in three re-circulating seawater tanks (4 inds./tank).
The fishes of three groups were intraperitoneally injected with 20-μL low-concentration bacterial suspension (1.0×107 colony forming units/mL; Infected 1), 20-μL high-concentration bacterial suspension (1.0×108 colony forming units/mL; Infected 2), and 20-μL phosphate buffer saline (PBS) (control group), respectively. All seahorse individuals were anesthetized with 0.035% Tricaine Methanesulfonate (MS-222) (Sigma-Aldrich, Castle Hill, NSW, Australia) and dissected rapidly at 24-h post-infection. The tissues were frozen in liquid nitrogen and stored at -80 ℃ until RNA extraction. We confirm that all experimental protocols were approved by the Animal Care and Use Committee of the Ludong University.
2.2 RNA isolation, cDNA synthesis, and real-time quantitative PCR analysisTotal RNA was extracted using TRIzol reagent (TaKaRa, Dalian, China) according to the manufacturer's instructions. The first-strand cDNA was synthesized using an oligo (dT) primer with the RNA Eraser Reverse Transcription Kit (TaKaRa, Japan). Specific PCR primers were designed to amplify these cDNA fragments by using Primer Premier 5.0 (Table 1). The real-time quantitative PCR (qPCR) was performed on Roche Light Cycler 480 using SYBR Premix Ex TaqTM (TaKaRa, Japan), and the PCR conditions were as follows: 40 cycles at 94 ℃ for 20 s, 52 ℃ for 20 s, and 72 ℃ for 15 s. Seahorse 18S rRNA was amplified to confirm the expression level of the housekeeping gene. Relative gene expression was determined using the 2-ΔΔCt comparative quantification method (Livak and Schmittgen, 2001).
RNA-seq data of H. erectus was downloaded from the NCBI Sequence Read Archive under accession number of SRA392578 (Lin et al., 2016). HTSeq v0.6.1 was used to count the numbers of reads mapped to each gene. Then, the expected number of Reads Per Kilobase of transcript sequence per Millions base pairs sequenced (RPKM) of each gene was calculated based on the length of the gene and read counts mapped to each gene.
2.4 Gene identification and sequence analysisThe whole genome sequence databases of the lined seahorse (H. erectus) were searched using the available teleosts' MAPK protein sequences, including those from Danio rerio, Poecilia formosa, Oryzias latipes, Larimichthys crocea, Oreochromis niloticus, Xiphophorus maculatus, and Cynoglossus semilaevis. TBLASTN algorithm was used to obtain the initial pool of MAPK family genes using a cut-off E-value of 10-5. All MAPK genes obtained from H. erectus genome were further confirmed against the published transcriptome sequences of H. erectus using the BLASTN program (Lin et al., 2017).
The exon/intron organization for MAPK genes were drawn by comparing the cDNA sequences with their corresponding genomic DNA sequences using the Gene Structure Display Server (GSDS) (http://gsds.cbi.pku.edu.cn). The protein domains were predicted by Simple Modular Architecture Research Tool (SMART) (http://smart.emblheidelberg.de).
2.5 Phylogenetic analysesAll MAPK protein sequences obtained from H. erectus, D. rerio, P. formosa, O. latipes, L. crocea, O. niloticus, X. maculatus, and C. semilaevis were aligned using MAFFT version 7 (Katoh and Standley, 2013). The best amino acid substitution model was chosen under the Akaike Information Criterion (AIC) as calculated in ProtTest (Abascal et al., 2005). Maximum-likelihood (ML) analysis was implemented in PhyML 3.0 with 1 000 bootstrap replicates (Guindon et al., 2010). The final ML tree was visualized using FigTree v1.4.2.
2.6 Selection analysesTo assess the rates of non-synonymous (Ka) and synonymous (Ks) substitutions for all MAPK codon sites and branches of the phylogeny of MAPKs, the branch-site model (BSM) and branch model (BM) were tested using the CODEML program of PAML v4.8 package (Yang, 2007). The BM was employed under two assumptions: one-ratio model and free-ratio model. For BSM, comparison between BSM A and the corresponding null model were constructed, and the codon sites under positive selection (Ka/Ks > 1) in the foreground branches was detected.
The likelihood ratio test (LRT) was constructed to verify that the best models fitted the data, and Bayes Empirical Bayes (BEB) approach was applied to identify amino acid codon sites under positive selection based on a posterior probability threshold of 0.95. The Ka/Ks ratios of the main branch of the phylogeny of MAPKs were labeled and the positive selected sites were displayed in a graphic view of sequence alignment using BioEdit v7.1.3.0.
2.7 Statistical analysesStatistical analyses were conducted using the programs SPSS 19.0 (IBM, US). The gene expression in different seahorse treatment groups were analyzed using the one-way analysis of variance (ANOVA).
3 RESULT 3.1 Gene identification and structure analysis of MAPK genes of H. erectusSeven MAPK genes, including erk1 (also known as mapk3), erk2 (mapk1), erk5 (mapk7), jnk1 (mapk8), jnk2 (mapk9), jnk3 (mapk10), and p38α (mapk14), were identified from the H. erectus genome, and the predicted sequences were further confirmed against the published transcriptome data of H. erectus. The open reading frames of erk1, erk2, erk5, jnk1, jnk2, jnk3, and p38α were 1 185, 1 857, 3 156, 1 695, 1 368, 1 536, and 1 137 bp, respectively, and encoded 394, 618, 1 051, 564, 455, 511, and 378 amino acids (Fig. 1). In addition, the genomic DNA sequences of each MAPK genes were obtained from the H. erectus genome based on the genome annotation file (gff3 format).
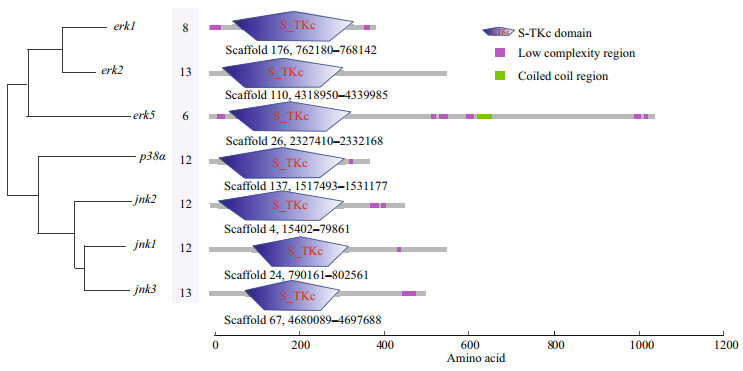
|
| Fig.1 The gene structures of MAPK genes in H. erectus genome The numbers indicate the exon number of corresponding genes, and the gene positions are listed below the gene structure. S-TKc: Serine/Threonine protein kinases, catalytic domain. |
The exon-intron junctions were determined by comparing the cDNA sequences with their corresponding genomic DNA sequences. We found that erk2 and jnk3 were organized into 13 exons and 12 introns, and jnk1, jnk2, p38α were composed of 12 exons. Moreover, the erk1 and erk5 were only composed of 8 and 6 exons, respectively. Some introns are extremely long (> 4 kb), including the fourth and fifth introns in erk2, the eleventh intron in p38α and jnk2, and the first intron in jnk3. The quantity of exons exhibited little difference among the erk2, jnk3, p38α, jnk1, and jnk2 (13, 13, 12, 12, and 12, respectively) (Fig. 1).
3.2 Phylogenetic analysis of MAPK genes of H. erectusIn this study, we constructed a phylogenetic tree of the 56 MAPK genes from 8 teleost species (Fig. 2). The ML tree shows that the MAPK genes formed three well-defined clades, corresponding to erk, jnk, and p38α MAPK subfamilies. As we predicted, each gene formed its own clade, which provided a solid phylogenetic evidence that was necessary to confirm the accuracy of identification of the H. erectus MAPKs. Our results show that erk1 and erk2 formed a sister group, and then clustered with erk5. The phylogeny suggested a sister relationship between jnk1 and jnk3, and then clustered with jnk2. The p38α was identified as the sister group of the jnk subfamily. Different H. erectus MAPK genes had a different clustering pattern when compared in gene homology with other teleosts. H. erectus was clustered with C. semilaevis in the clades of erk1 and erk5, while in the phylogeny of jnk1 and jnk3, H. erectus was clustered with O. latipes and D. rerio.
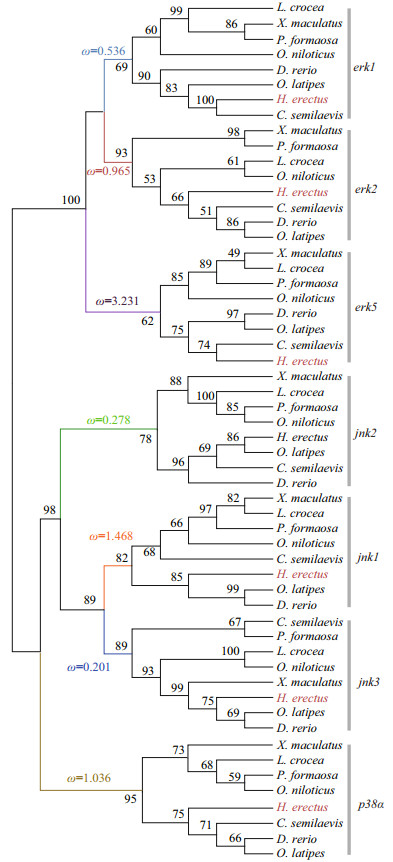
|
| Fig.2 Phylogenetic tree and selection pressure of MAPK genes from the lined seahorse (H. erectus) and other fish |
Selective pressure of different MAPK subfamilies after gene duplication were detected. The Ka/Ks values were calculated across all branches using the branch model of CODEML. The results suggest that there was a substantial difference in selective pressure among the MAPK subfamilies. Signatures of positive selection in erk5 (Ka/Ks=3.231), jnk1 (Ka/Ks=1.468), p38α (Ka/Ks=1.036) were revealed in the phylogeny, which may suggested that adaptive changes in these genes (Fig. 2). The Ka/Ks values estimated in erk1, erk2, jnk2, and jnk3 were less than 1 (0.536, 0.965, 0.278, and 0.201, respectively).
3.3 Positive selection analysis of MAPK genes of H. erectusThe branch-site model in CODEML was used to reveal whether Hippocampus MAPK genes were subjected to positive selection. We set the H. erectus and H. comes as a foreground branch, the specific codons identified by the BEB approach with a posterior probability of 95% are listed in the Table 2. We found that four positive selection sites among four MAPK genes of seahorse that showed BEB values > 0.95, including erk2, erk5, jnk1, and p38α, which may have a plasticity in function (Fig. 3, Table 2). The 191T in erk2 and the 351A in erk5 were identified as positive selection sites with a BEB posterior probability of 96.6% and 98.3%, respectively. Positively selected sites located in the amino acid positions 349 and 45 were detected in the jnk1 and p38α with a posterior probability of 98.1% and 95.9%.
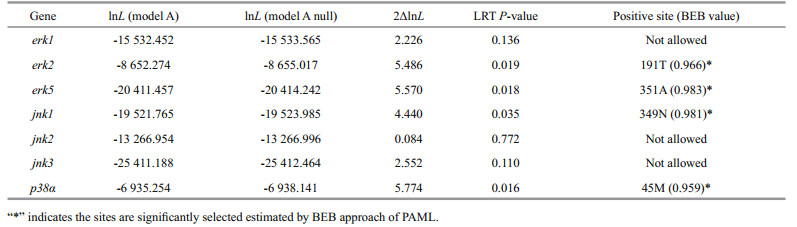
|
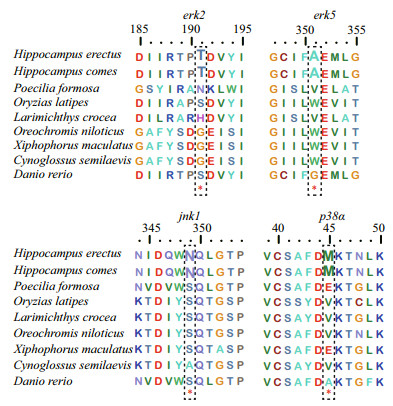
|
| Fig.3 Positive selection sites detected in MAPK family genes |
In total, 34 genes involved in the MAPK cascade signal transduction pathway were identified from the RNA-seq data, and most genes have shown different expression trends between different tissues and development stages (Fig. 4). A stage-specific gene expression profile was revealed from the comparative transcriptome analysis, and low expression level of many genes were detected in the testis and brood pouch at the age of 5 months, including raf (raf proto oncogene serine/threonine protein kinase), mlk2 (mixed lineage kinase 2), mlk3, mekk4 (mitogen-activated protein kinase kinase kinase 4), tao1 (serine/threonine-protein kinase tao1), tao2, mkk2 (mitogen-activated protein kinase kinase 2), mkk7, erk1, msk2 (ribosomal protein s6 kinase alpha-4), etc. In the first 30 days after birth, a novel expression profile of MAPK cascade genes was found during the first day after birth. Compared with other develop stages, the expression level of many genes were lowest in the 1-day-old larvae, including tao1, tao2, ask1 (apoptosis signal-regulating kinase 1), mkk1, mkk4, mkk5, mkk6, p38α, jnk3, rsk2 (ribosomal s6 kinase 2), rsk3, msk1, mk2, and mk3. However, the genes including raf, mos, tak1, mkk2, erk1, p38β, jnk2, and mnk1 showed highest expression level in the 1-day-old larvae (Fig. 4).
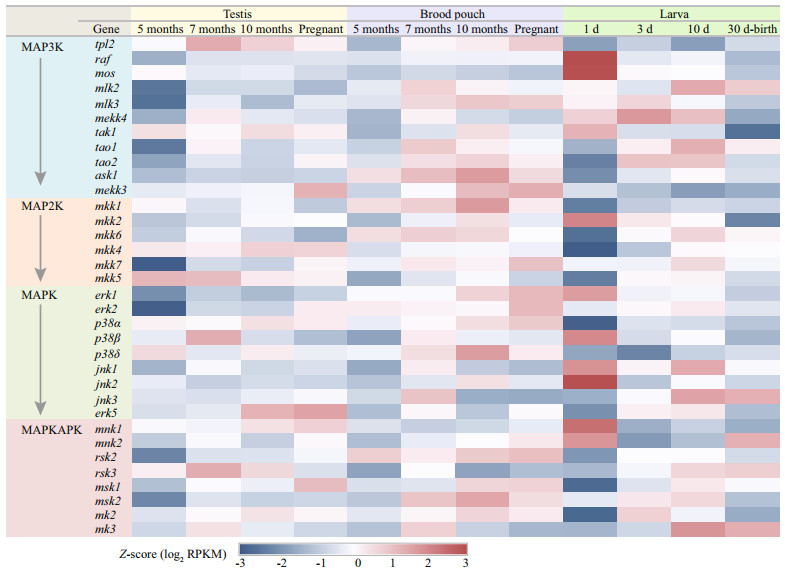
|
| Fig.4 Transcriptomics data analysis of the lined seahorse Z-scores were calculated from RPKM values. |
The expressions levels of the four positively selected MAPK genes together with jnk2, jnk3 were analyzed in nine tissues of the H. erectus (Fig. 5). The result show the expression of the mRNA of erk2 and jnk2 gene was highest in intestine, the mRNA of erk5, jnk1, and p38α were all most abundantly expressed in the ovary, and the gene expression of jnk3 was highest in gill.
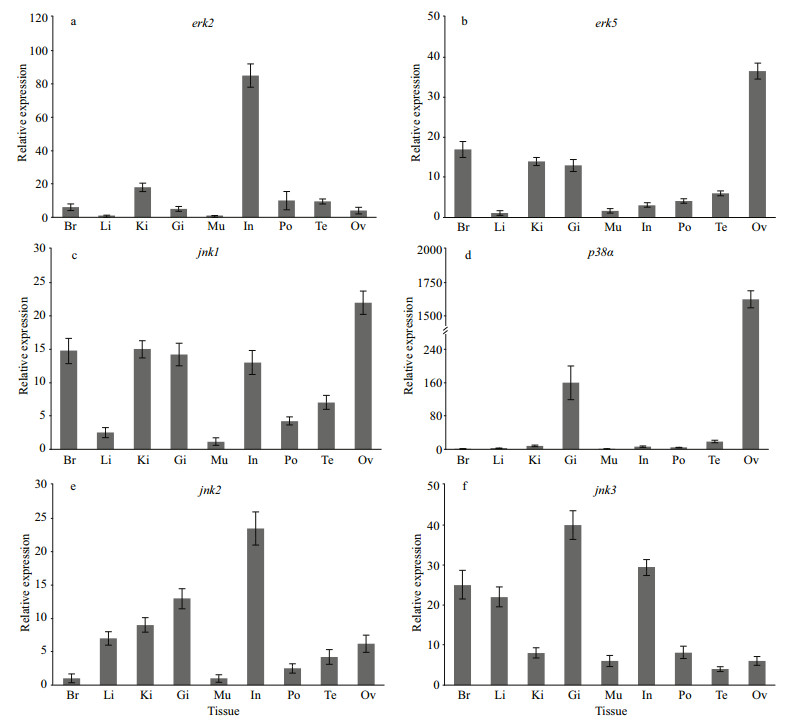
|
| Fig.5 The expression levels of MAPK family genes in different tissues Br: brain; Li: liver; Ki: kidney; Gi: gill; Mu: muscle; In: intestine; Po: brood pouch; Te: testis; Ov: Ovary. mRNA levels are expressed as a ratio relative to the 18S rRNA levels. |
Challenge of the lined seahorse with the V. fortis resulted in a significant up-regulation of the erk2, erk5, jnk1, jnk2, jnk3, and p38α genes in different tissues (Fig. 6). The mRNA expression level of erk2, erk5, and jnk3 in the liver were significantly increased after injection with the pathogenic bacteria V. fortis. In the gill, the transcript of erk2, erk5, jnk1, jnk2, jnk3, and p38α were significantly increased after experimental challenge. However, the relative level of expression of the four genes tested in ovary and testis does not change significantly post-infection, except for the jnk1. The mRNA expression level of erk2, jnk1, jnk2, and p38α were significantly increased after challenge in the intestine. Moreover, in the brood pouch, the mRNA expression level of erk2, erk5, and jnk1 were significantly increased after challenge, and expression of these three genes does not change significantly following the injection of low and high concentrations of V. fortis (Fig. 6).
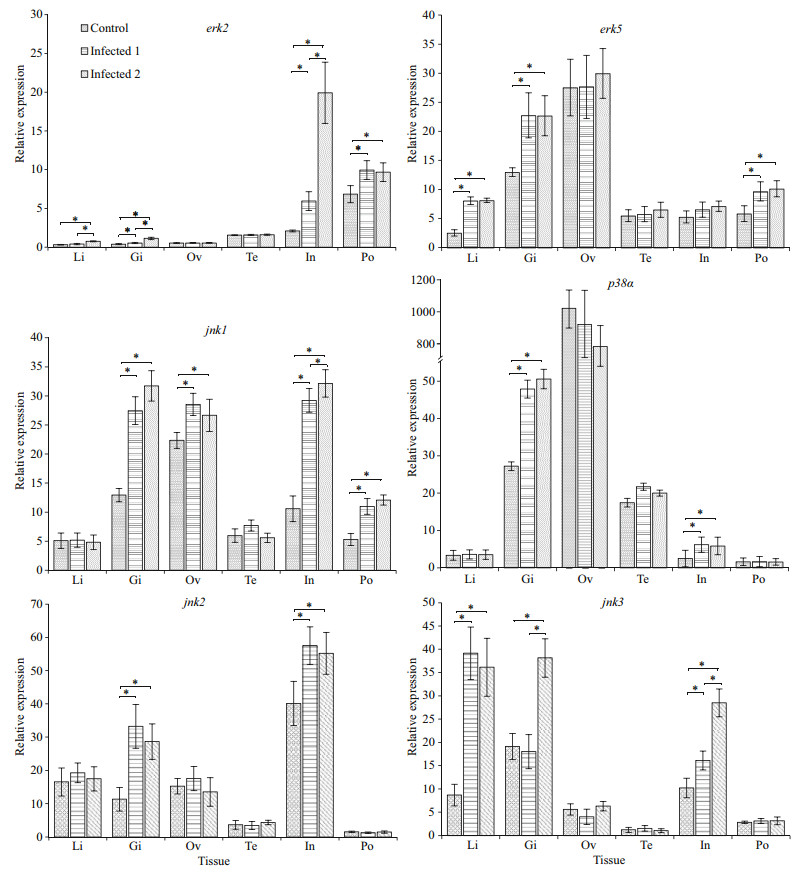
|
| Fig.6 Relative expression of the MAPK genes in various tissues of the healthy lined seahorses and V. fortis challenged lined seahorses mRNA levels are expressed as a ratio relative to the 18S rRNA levels in the same samples after qPCR. *: represents significance at P < 0.05. Li: liver; Gi: gill; Ov: Ovary; Te: testis; In: intestine; Po: brood pouch. |
Previous genome-wide studies of the seahorse have indicated a substantial loss of conserved noncoding elements, which were indicated that some peculiar physiological mechanism in metabolism and regulation is developed in seahorse (Lin et al., 2016). MAPK genes are important transcription factors that convert extracellular stimuli into a wide range of fundamental cellular responses. Although the genomic structures of H. erectus MAPK genes are varied, the evolutionarily conserved serine/threonine protein kinase catalytic (S-TKc) domain was found in all MAPK proteins, which were similar to those found in other species (Jia et al., 2015). The gene structure of MAPK genes suggested the high conservation in gene splicing during the evolutionary history. The specific occurrence of intron losses in the erk1 and erk7 subfamilies might be associated with adaptive evolution of the teleosts by enhancing the gene expression level of both MAPK genes (Kawaguchi et al., 2010; Li et al., 2011). The evolution of MAPK gene family is highly conserved throughout the eukaryotic kingdoms (Cook et al., 1997; Hamel et al., 2012). However, the detailed evolutionary history of the vertebrate MAPK family is largely unclear (Li et al., 2011). Gene duplications of the MAPK family genes were found to be ubiquitous in the evolutionary history, and the whole-genome duplication (WGD) is believed to be a really important source of evolutionary changes. Additionally, the redundant genes derived from WGD are considered to be acquire new or altered functions (Sémon and Wolfe, 2007; Inoue et al., 2015). The evolutionary history of teleost MAPK family needs to be confirmed by a systematic phylogenetic analysis. We found that these teleosts were conventional in MAPK family gene members, and the MAPK family had been formed through multiple duplications at least prior to the diversification of teleost fishes (Li et al., 2011). The deep divergence of MAPK genes may have benefited from the extra event of whole-genome duplication in teleost fish, which is proposed to have provided sufficient genetic material for the adaptive evolution (Toloza-Villalobos et al., 2015).
Whether the natural selection has driven the evolution of the teleost MAPK family remains unknown, and it will be interesting to find compelling evidence of positive selections acting of MAPK genes in H. erectus and H. comes. The MAPK family genes have been considered important in the adaptive evolution of the vertebrates (Li et al., 2011). The selection results based on the branch model indicating that purifying selection dominated the evolution of these genes with strong functional constraints. Previous study suggested that MAPK genes have high level of selective constraints are under strong functional constraints, although there is an expansion of MAPK gene family (Wu et al., 2010; Li et al., 2011). However, different H. erectus MAPK genes have been under different selective pressures. The positive selection of erk2, erk5, jnk1, and p38α observed in the present study could help us to better understand the evolutionary history of seahorses and its adaptation to the complex and changeable offshore environment. The specific gene expression pattern and the positive selection detected in the corresponding gene of H. erectus may reflected a plasticity in gene function, which will facilitated the radiation of the group.
The innovation of "male pregnancy" in seahorses and pipefishes, together with novel expression patterns of key immune and pregnancy genes in their reproductive tissues, were considered to be an important research system for studying vertebrate immune system (Roth et al., 2020). A problem for pregnancy to evolve is the rejection of the embryo that is recognized as foreign tissue by the vertebrate's immune system (La Rocca et al., 2014; Roth et al., 2020). In the present study, a novel expression profile of many MAPK cascade genes was found in the seahorse larvae during the first day after birth, which may reflects a signs of immune response to its parental immune system. The brood pouch, where the entire embryonic development takes place, has shown significant transcriptional changes in response to pathogen infection as reported by Roth et al. (2020). However, as reproductive organs, the ovaries, and testes did not respond sufficiently to external infections, which may indicates that the brood pouch is an important immune barrier in seahorse. In addition, the results suggest that liver and gill, the important immune organs in teleost fish, may play an important role in immune protection. The seahorse has a siphon-like snout and abandoned predatory life, which may result a less injury and infection in its gastrointestinal tract (Luo et al., 2016). Previous studies have confirmed that the seahorse lacks a spleen and GALT (Galtier et al., 2009; Litman et al., 2010; Luo et al., 2016), and we found that the intestinal has a significant upregulation response to bacterial infection, which may be attributed to the intestinal immune compensation to "immunodeficiency" of the seahorse. Because the seahorses are well known for its breeding strategy and novel immunological characters, the adaptive change in their MAPK genes with vital immune functions may provide clues for the studies of breeding adaptation in seahorses.
5 CONCLUSIONSeven MAPK subfamilies including erk1, erk2, erk5, jnk1, jnk2, jnk3, and p38α were successfully identified from the lined seahorse Hippocampus erectus. We characterized their gene structure and protein domains, and found intron losses in the erk1 and erk7 subfamilies. Phylogeny of the MAPK family genes suggested multiple duplication events prior to the diversification of teleosts were occurred. Selective pressure of different MAPK subfamilies indicated the adaptive changes in erk5, jnk1, and p38α, and positive selection sites were found in erk2, erk5, jnk1, and p38α. The expression pattern of jnk2, jnk3, and four positively selected MAPK genes were analyzed in nine tissues of the H. erectus, among which erk5, jnk1, and p38α were most abundantly expressed in the ovary. A novel expression profile of MAPK cascade genes was found in the seahorse larvae during the first day after birth, which reflected vital signs of immune response to its parental immune system. The expression patterns of these MAPK genes were analyzed following the bacterial challenge of V. fortis, revealing their upregulation pattern in brood pouch and other immune tissues. This study will enrich our knowledge of the evolution of the H. erectus MAPK subfamilies, and facilitate better understanding of the functional role of MAPKs in teleosts.
6 DATA AVAILABILITY STATEMENTData that support the findings of this study are available from the corresponding author upon reasonable request.
Abascal F, Zardoya R, Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics, 21(9): 2104-2105.
DOI:10.1093/bioinformatics/bti263 |
Antonopoulou E, Kentepozidou E, Feidantsis K, Roufidou C, Despoti S, Chatzifotis S. 2013. Starvation and re-feeding affect Hsp expression, MAPK activation and antioxidant enzymes activity of European sea bass (Dicentrarchus labrax). Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology, 165(1): 79-88.
DOI:10.1016/j.cbpa.2013.02.019 |
Arthur J S C, Ley S C. 2013. Mitogen-activated protein kinases in innate immunity. Nature reviews Immunology, 13(9): 679-692.
DOI:10.1038/nri3495 |
Cargnello M, Roux P P. 2011. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiology and Molecular Biology Reviews, 75(1): 50-83.
DOI:10.1128/MMBR.00031-10 |
Codina M, García de la serrana D, Sánchez-Gurmaches J, Montserrat N, Chistyakova O, Navarro I, Gutiérrez J. 2008. Metabolic and mitogenic effects of IGF-Ⅱ in rainbow trout (Oncorhynchus mykiss) myocytes in culture and the role of IGF-Ⅱ in the PI3K/Akt and MAPK signalling pathways. General and Comparative Endocrinology, 157(2): 116-124.
DOI:10.1016/j.ygcen.2008.04.009 |
Cook J G, Bardwell L, Thorner J. 1997. Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous-growth signalling pathway. Nature, 390(6655): 85-88.
DOI:10.1038/36355 |
Coulombe P, Meloche S. 2007. Atypical mitogen-activated protein kinases: structure, regulation and functions. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research, 1773(8): 1376-1387.
DOI:10.1016/j.bbamcr.2006.11.001 |
Ding X, Liang Y S, Peng W, Li R Z, Lin H R, Zhang Y, Lu D Q. 2018. Intracellular TLR22 acts as an inflammation equalizer via suppression of NF-κB and selective activation of MAPK pathway in fish. Fish & Shellfish Immunology, 72: 646-657.
DOI:10.1016/j.fsi.2017.11.042 |
Feidantsis K, Pörtner H O, Markou T, Lazou A, Michaelidis B. 2012. Involvement of p38 MAPK in the induction of Hsp70 during acute thermal stress in red blood cells of the gilthead sea bream, Sparus aurata. Journal of Experimental Zoology Part A—Ecological Genetics and Physiology, 317(5): 303-310.
DOI:10.1002/jez.1725 |
Foster S J, Vincent A C J. 2004. Life history and ecology of seahorses: implications for conservation and management. Journal of Fish Biology, 65(1): 1-61.
DOI:10.1111/j.0022-1112.2004.00429.x |
Fuentes E N, Björnsson B T, Valdés J A, Einarsdottir I E, Lorca B, Alvarez M, Molina A. 2011. IGF-I/PI3K/Akt and IGF-I/MAPK/ERK pathways in vivo in skeletal muscle are regulated by nutrition and contribute to somatic growth in the fine flounder. American Journal of Physiology, Regulatory, Integrative and Comparative Physiology, 300(6): 1532-1542.
DOI:10.1152/ajpregu.00535.2010 |
Galtier N, Nabholz B, Glémin S, Hurst G D D. 2009. Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Molecular Ecology, 18(22): 4541-4550.
DOI:10.1111/j.1365-294X.2009.04380.x |
Guindon S, Dufayard J F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology, 59(3): 307-321.
DOI:10.1093/sysbio/syq010 |
Hamel L P, Nicole M C, Duplessis S, Ellis B E. 2012. Mitogen-activated protein kinase signaling in plant-interacting fungi: distinct messages from conserved messengers. Plant Cell, 24(4): 1327-1351.
DOI:10.1105/tpc.112.096156 |
He Y, Cai C, Sun S, Wang X, Li W, Li H. 2016. Effect of JNK inhibitor SP600125 on hair cell regeneration in zebrafish (Danio rerio) larvae. Oncotarget, 7: 51640-51650.
DOI:10.18632/oncotarget.10540 |
Iliev D B, Hansen T, Jørgensen S M, Krasnov A, Jørgensen J B. 2013. CpG- and LPS-activated MAPK signaling in in vitro cultured salmon (Salmo salar) mononuclear phagocytes. Fish & Shellfish Immunology, 35(4): 1079-1085.
DOI:10.1016/j.fsi.2013.07.014 |
Inoue J, Sato Y, Sinclair R, Tsukamoto K, Nishida M. 2015. Rapid genome reshaping by multiple-gene loss after whole-genome duplication in teleost fish suggested by mathematical modeling. Proceedings of the National Academy of Sciences of the United States of America, 112(48): 14918-14923.
DOI:10.1073/pnas.1507669112 |
Jia Q J, Fan Z J, Yao C L. 2015. Identification and expression profiles of erk2 and erk5 in large yellow croaker (Larimichthys crocea) after temperature stress and immune challenge. Fish & Shellfish Immunology, 44(2): 410-419.
DOI:10.1016/j.fsi.2015.03.006 |
Jopling C, Suñè G, Morera C, Izpisua Belmote J C. 2012. p38α MAPK regulates myocardial regeneration in zebrafish. Cell Cycle, 11(6): 1195-1201.
DOI:10.4161/cc.11.6.19637 |
Katoh K, Standley D M. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution, 30(4): 772-780.
DOI:10.1093/molbev/mst010 |
Kawaguchi, M, Hiroi, J, Mi ya, M, Nishida, M, Iuchi, I, Yasumasu, S. 2010. Intron-loss evolution of hatching enzyme genes in Teleostei. BMC Evolutionary Biology, 10: 260.
DOI:10.1186/1471-2148-10-260 |
Keller J M, Escara-Wilke J F, Keller E T. 2008. Heat stress-induced heat shock protein 70 expression is dependent on ERK activation in zebrafish (Danio rerio) cells. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology, 150(3): 307-314.
DOI:10.1016/j.cbpa.2008.03.021 |
Krens S F G, He S N, Lamers G E M, Meijer A H, Bakkers J, Schmidt T, Spaink H P, Snaar-Jagalska B E. 2008. Distinct functions for ERK1 and erk2 in cell migration processes during zebrafish gastrulation. Developmental Biology, 319(2): 370-383.
DOI:10.1016/j.ydbio.2008.04.032 |
Krens S F G, He S N, Spaink H P, Snaar-Jagalska B E. 2006. Characterization and expression patterns of the MAPK family in zebrafish. Gene Expression Patterns, 6(8): 1019-1026.
DOI:10.1016/j.modgep.2006.04.008 |
Kültz D, Avila K. 2001. Mitogen-activated protein kinases are in vivo transducers of osmosensory signals in fish gill cells. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology, 129(4): 821-829.
DOI:10.1016/s1096-4959(01)00395-5 |
Kyriakis J M, Avruch J. 2012. Mammalian MAPK signal transduction pathways activated by stress and inflammation: a 10-year update. Physiological Reviews, 92(2): 689-737.
DOI:10.1152/physrev.00028.2011 |
La Rocca C, Carbone F, Longobardi S, Matarese G. 2014. The immunology of pregnancy: regulatory T cells control maternal immune tolerance toward the fetus. Immunology Letters, 162(1): 41-48.
DOI:10.1016/j.imlet.2014.06.013 |
Leal R B, Ribeiro S J, Posser T, Cordova F M, Rigon A P, Filho E Z, Bainy A C D. 2006. Modulation of ERK1/2 and p38MAPK by lead in the cerebellum of Brazilian catfish Rhamdia quelen. Aquatic Toxicology, 77(1): 98-104.
DOI:10.1016/j.aquatox.2005.11.002 |
Li M, Liu J, Zhang C Y. 2011. Evolutionary history of the vertebrate mitogen activated protein kinases family. PLoS One, 6(10): e26999.
DOI:10.1371/journal.pone.0026999 |
Li X Y, Ma J G, Li Y Y. 2016. Molecular cloning and expression determination of p38 MAPK from the liver and kidney of silver carp. Journal of Biochemical and Molecular Toxicology, 30(5): 224-231.
DOI:10.1002/jbt.21781 |
Lin Q, Fan S H, Zhang Y H, Xu M, Zhang H X, Yang Y L, Lee A P, Woltering J M, Ravi V, Gunter H M, Luo W, Gao Z X, Lim Z W, Qin G, Schneider R F, Wang X, Xiong P W, Li G, Wang K, Min J M, Zhang C, Qiu Y, Bai J, He W M, Bian C, Zhang X H, Shan D, Qu H Y, Sun Y, Gao Q, Huang L M, Shi Q, Meyer A, Venkatesh B. 2016. The seahorse genome and the evolution of its specialized morphology. Nature, 540(7633): 395-399.
DOI:10.1038/nature20595 |
Lin Q, Qiu Y, Gu R B, Xu M, Li J, Bian C, Zhang H X, Qin G, Zhang Y H, Luo W, Chen J M, You X X, Fan M J, Sun M, Xu P, Venkatesh B, Xu J M, Fu H T, Shi Q. 2017. Draft genome of the lined seahorse, Hippocampus erectus. Giga Science, 6(6): 1-6.
DOI:10.1093/gigascience/gix030 |
Litman G W, Rast J P, Fugmann S D. 2010. The origins of vertebrate adaptive immunity. Nature Reviews Immunology, 10(8): 543-553.
DOI:10.1038/nri2807 |
Livak K J, Schmittgen T D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods, 25(4): 402-408.
DOI:10.1006/meth.2001.1262 |
Luo W, Wang X, Qu H Y, Qin G, Zhang H X, Lin Q. 2016. Genomic structure and expression pattern of MHC Ⅱα and Ⅱ β genes reveal an unusual immune trait in lined seahorse Hippocampus erectus. Fish & Shellfish Immunology, 58: 521-529.
DOI:10.1016/j.fsi.2016.09.057 |
Marshall W S, Cozzi R R F, Spieker M. 2017. WNK1 and p38-MAPK distribution in ionocytes and accessory cells of euryhaline teleost fish implies ionoregulatory function. Biology Open, 6(7): 956-966.
DOI:10.1242/bio.024232 |
Marshall W S, Ossum C G, Hoffmann E K. 2005. Hypotonic shock mediation by p38 MAPK, JNK, PKC, FAK, OSR1 and SPAK in osmosensing chloride secreting cells of killifish opercular epithelium. Journal of Experimental Biology, 208(Pt 6): 1063-1077.
DOI:10.1242/jeb.01491 |
Matsunaga T, Rahman A. 1998. What brought the adaptive immune system to vertebrates?—The jaw hypothesis and the seahorse. Immunological Reviews, 166(1): 177-186.
DOI:10.1111/j.1600-065x.1998.tb01262.x |
Olavarria V H, Valdivia S, Salas B, Villalba M, Sandoval R, Oliva H, Valdebenito S, Yañez A. 2015. ISA virus regulates the generation of reactive oxygen species and p47 phox expression in a p38 MAPK-dependent manner in Salmo salar. Molecular Immunology, 63(2): 227-234.
DOI:10.1016/j.molimm.2014.07.016 |
Plotnikov A, Zehorai E, Procaccia S, Seger R. 2011. The MAPK cascades: signaling components, nuclear roles and mechanisms of nuclear translocation. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research, 1813(9): 1619-1633.
DOI:10.1016/j.bbamcr.2010.12.012 |
Qin G, Zhang Y H, Ho A L F C, Zhang Y, Lin Q. 2017. Seasonal distribution and reproductive strategy of seahorses. ICES Journal of Marine Science, 74(8): 2170-2179.
DOI:10.1093/icesjms/fsx042 |
Raman M, Chen W, Cobb M H. 2007. Differential regulation and properties of MAPKs. Oncogene, 26(22): 3100-3112.
DOI:10.1038/sj.onc.1210392 |
Roth O, Solbakken M H, Tørresen O K, Bayer T, Matschiner M, Baalsrud H T, Hoff S N K, Brieuc M S O, Haase D, Hanel R, Reusch T B H, Jentoft S. 2020. Evolution of male pregnancy associated with remodeling of canonical vertebrate immunity in seahorses and pipefishes. Proceedings of the National Academy of Sciences of the United States of America, 117(17): 9431-9439.
DOI:10.1073/pnas.1916251117 |
Sánchez-Gurmaches J, Cruz-Garcia L, Ibarz A, Fernández-Borrás J, Blasco J, Gutiérrez J, Navarro I. 2013. Insulin, IGF-I, and muscle MAPK pathway responses after sustained exercise and their contribution to growth and lipid metabolism regulation in gilthead sea bream. Domestic Animal Endocrinology, 45(3): 145-153.
DOI:10.1016/j.domaniend.2013.08.001 |
Sémon M, Wolfe K H. 2007. Rearrangement rate following the whole-genome duplication in teleosts. Molecular Biology and Evolution, 24(3): 860-867.
DOI:10.1093/molbev/msm003 |
Sopontammarak S, Aliharoob A, Ocampo C, Arcilla R A, Gupta M P, Gupta M. 2005. Mitogen-activated protein kinases (p38 and c-Jun NH2-terminal kinase) are differentially regulated during cardiac volume and pressure overload hypertrophy. Cell Biochemistry and Biophysics, 43(1): 61-76.
DOI:10.1385/CBB:43:1:061 |
Sun Y, Zhang L L, Zhang M W, Li R J, Li Y P, Hu X L, Wang S, Bao Z M. 2016. Characterization of three mitogen-activated protein kinases (MAPK) genes reveals involvement of ERK and JNK, not p38 in defense against bacterial infection in Yesso scallop Patinopecten yessoensis. Fish & Shellfish Immunology, 54: 507-515.
DOI:10.1016/j.fsi.2016.04.139 |
Toloza-Villalobos J, Ignacio Arroyo J, Opazo J C. 2015. The circadian clock of teleost fish: a comparative analysis reveals distinct fates for duplicated genes. Journal of Molecular Evolution, 80(1): 57-64.
DOI:10.1007/s00239-014-9660-x |
Urushibara N, Mitsuhashi S, Sasaki T, Kasai H, Yoshimizu M, Fujita H, Oda A. 2009. JNK and p38 MAPK are independently involved in tributyltin-mediated cell death in rainbow trout (Oncorhynchus mykiss) RTG-2 cells. Comparative Biochemistry and Physiology Part C: Toxicology & Pharmacology, 149(4): 468-475.
DOI:10.1016/j.cbpc.2008.10.109 |
Wang X, Han X, Zhang Y, Liu S S, Lin Q. 2019. Phylogenetic analysis and genetic structure of the seahorse, Hippocampus fuscus from theArabian and Red Sea based on mitochondrial DNA sequences. Mitochondrial DNA Part A: DNA Mapping, Sequencing, and Analysis, 30(1): 165-171.
DOI:10.1080/24701394.2018.1467410 |
Wu X C, Chi X Q, Wang P M, Zheng D Q, Ding R, Li Y D. 2010. The evolutionary rate variation among genes of HOG-signaling pathway in yeast genomes. Biology Direct, 5: 46.
DOI:10.1186/1745-6150-5-46 |
Yang Z H. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution, 24(8): 1586-1591.
DOI:10.1093/molbev/msm088 |
Zhang Y H, Qin G, Zhang H X, Wang X, Lin Q. 2017. DNA barcoding reflects the diversity and variety of brooding traits of fish species in the family Syngnathidae along China's coast. Fisheries Research, 185: 137-144.
DOI:10.1016/j.fishres.2016.09.015 |
Zhu J, Cai L, Zhang T H, Chen L M, Jin P, Ma F. 2014. Identification and characterization of a p38-like gene from amphioxus (Branchiostoma belcheri): an insight into amphioxus innate immunity and evolution. Fish & Shellfish Immunology, 41(2): 421-427.
DOI:10.1016/j.fsi.2014.09.028 |
 2021, Vol. 39
2021, Vol. 39



