Institute of Oceanology, Chinese Academy of Sciences
Article Information
- DU Haijian, ZHANG Wenyan, LIN Wei, PAN Hongmiao, XIAO Tian, WU Long-Fei
- Genomic analysis of a pure culture of magnetotactic bacterium Terasakiella sp. SH-1
- Journal of Oceanology and Limnology, 39(6): 2097-2106
- http://dx.doi.org/10.1007/s00343-021-1054-5
Article History
- Received Feb. 10, 2021
- accepted in principle Mar. 11, 2021
- accepted for publication Apr. 22, 2021
2 CAS Key Laboratory of Marine Ecology and Environmental Sciences, Institute of Oceanology, Chinese Academy of Sciences, Qingdao 266071, China;
3 Laboratory for Marine Ecology and Environmental Science, Pilot National Laboratory for Marine Science and Technology(Qingdao), Qingdao 266237, China;
4 Center for Ocean Mega-Science, Chinese Academy of Sciences, Qingdao 266071, China;
5 Key Laboratory of Earth and Planetary Physics, Institute of Geology and Geophysics, Chinese Academy of Sciences, Beijing 100029, China;
6 Institutions of Earth Science, Chinese Academy of Sciences, Beijing 100029, China;
7 France-China Joint Laboratory for Evolution and Development of Magnetotactic Multicellular Organisms, Chinese Academy of Sciences, Beijing 100029, China;
8 Aix-Marseille University, CNRS, LCB, Marseille F-13402, France
Magnetotactic bacteria (MTB) are prokaryotes that orient and migrate along the geomagnetic field lines, a behavior referred to as magnetotaxis or microbial magnetoreception. MTB share the capacity to synthesize magnetosomes, which are magnetic crystals of magnetite (Fe3O4) and/or greigite (Fe3S4) and enveloped by a phospholipid bilayer membrane (Blakemore, 1982; Bazylinski et al., 1995; Bazylinski and Frankel, 2004). Many morphotypes of MTB, including coccoid, ovoid, rod, vibrio, spirillum, and multicellular forms have been observed worldwide across from freshwater, brackish, and marine waters to waterlogged soils (Maratea and Blakemore, 1981; Hanzlik et al., 2002; Sakaguchi et al., 2002; Lefèvre et al., 2009; Wenter et al., 2009; Zhou et al., 2013; Lin and Pan, 2015; Lin et al., 2017a).
All known MTB belong to the domain Bacteria, predominantly within the phylum Proteobacteria (Simmons et al., 2004; Jogler and Schüler, 2009; Lefèvre and Bazylinski, 2013; Ji et al., 2017; Lin et al., 2018). A diverse group of MTB affiliated with the phylum Nitrospirae, the candidate phylum Omnitrophica, the candidate phylum Latescibacteria, and the phylum Planctomycetes have been identified (Vali et al., 1987; Lefèvre et al., 2010; Kolinko et al., 2012; Lin et al., 2018). More recently, our knowledge of the phylogenetic diversity of MTB has been dramatically expanded and so far, these organisms have been found in up to 16 bacterial phylum-level lineages (Lin et al., 2020; Uzun et al., 2020). Most of the cultured MTB belong to the phylum Proteobacteria, and a large number of these are within the Alphaproteobacteria, such as freshwater Magnetospirillum species MS-1, AMB-1, MSR-1, and XM-1 (Blakemore et al., 1979; Matsunaga et al., 1991; Schleifer et al., 1991; Wang et al., 2016), marine vibrio strains MV-1 and MV-2 (Delong et al., 1993; Bazylinski et al., 2013), marine spirillum strains MMS-1 and QH-2 (Zhu et al., 2010; Williams et al., 2012), and recently isolated marine spirillum Candidatus Terasakiella magnetica strain PR-1 belonging to the genus Terasakiella (Monteil et al., 2018).
Much genomic data on MTB has now been obtained and analyzed. The whole genomes of some cultured MTB (e.g., AMB-1, MSR-1, QH-2, MC-1, MO-1, XM-1, BW-2, and SS-5) have been obtained (Matsunaga et al., 2005; Richter et al., 2007; Schübbe et al., 2009; Ji et al., 2014; Wang et al., 2016; Ji et al., 2017; Uebe et al., 2018; Geurink et al., 2020; Trubitsyn et al., 2021). While many other MTB remain to be draft sequenced. After decades of research on MTB genomes, many interesting features have been revealed. In particular amongst these is the presence of clustered genes that control magnetosome biomineralization and arrangement in MTB cells, which are termed magnetosome gene islands (MAIs) (Grünberg et al., 2001; Murat et al., 2010; Lohße et al., 2011) or magnetosome gene clusters (MGCs) (Lin et al., 2017b).
We recently isolated a new marine magnetotactic spirillum (designated Terasakiella sp. SH-1) into axenic culture from an intertidal zone in Sanya, China (Du et al., 2019). SH-1 belongs to the genus Terasakiella in the Alphaproteobacteria and is closely related to Ca. Terasakiella magnetica strain PR-1 (Monteil et al., 2018). In the previous study, we have reported a duplication event of magnetosome genes within mamAB operon in the genome of SH-1, which suggests that gene duplication event plays a potentially important role in the evolution of magnetotaxis in the Alphaproteobacteria (Du et al., 2019). Here, we perform a comparative analysis of the complete genome of SH-1 with representative MTB genomes, which provides novel insights in to metabolic potential and ecosystem function of this newly isolated MTB strains.
2 MATERIAL AND METHOD 2.1 Genome analysisThe whole-genome sequencing of the SH-1 was performed as previously described (Du et al., 2019). All genome data used in this study was downloaded from the NCBI site. The gene prediction was performed on the MicroScope platform (Vallenet et al., 2013). The tandem repeats annotation was obtained using the Tandem Repeat Finder (http://tandem.bu.edu/trf/trf.html) (Benson, 1999), and the minisatellite DNA and microsatellite DNA were selected based on the number and length of repeat units. Prophage regions were predicted using the PHAge Search Tool Enhanced Release (PHASTER) web server (http://phaster.ca/) (Arndt et al., 2016) and CRISPR identification using CRISPRFinder (Grissa et al., 2007). The best hit was performed using the BLAST (Altschul et al., 1990) alignment tool for function annotation. Seven databases including Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa et al., 2016), Clusters of Orthologous Groups (COG) (Galperin et al., 2015; Makarova et al., 2015), Non-Redundant Protein Database databases (NR), Swiss-Prot (Consortium, 2015), Gene Ontology (GO) (Ashburner et al., 2000), TrEMBL (Apweiler et al., 2004), and EggNOG (Huerta-Cepas et al., 2016) were used for general function annotation.
2.2 Nitrogen metabolism analysisProtein sequence similarities in relation to nitrogen metabolism were determined using BLAST on the MicroScope (Vallenet et al., 2013) platforms. Proteins in different organisms were defined as orthologs when their alignments met the criteria: E-value < 1e-5, identity > 30%, and query coverage > 50%. The gene markers used to decide whether particular transformation reactions were present were based on a published review (Kuypers et al., 2018) (Supplementary Table S1); the markers were for genes involved in 15 reactions involving eight key inorganic nitrogen species having different oxidation states.
2.3 Pan/core genome analysisA pan/core genome analysis using the MicroScope (Vallenet et al., 2013) gene families (MICFAM), computed using the SiLiX software (Miele et al., 2011) (threshold: 50% of amino-acid identity and 80% of alignment coverage).
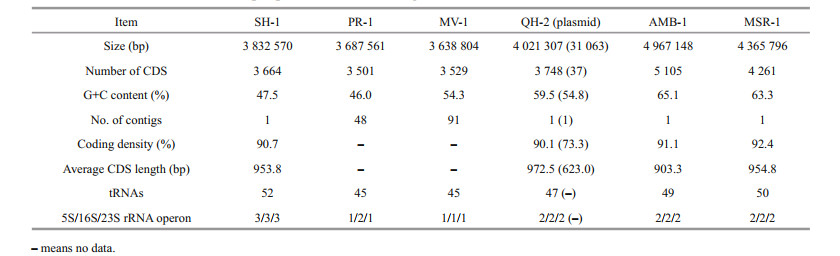
3 RESULT AND DISCUSSION 3.1 Genome overview of SH-1The genome of SH-1 comprised 3 832 570 bp in a circular chromosome having the average G+C content of 47.5%. The chromosome contained 3 664 predicted coding sequences (CDS), including those encoding 52 tRNAs and three sets of rRNAs (5S, 16S, and 23S), which corresponded to 90.7% of the genome being coding sequences. Compared to freshwater M. magneticum AMB-1 and M. gryphiswaldense MSR-1, the genome size, G+C content, and CDS number of marine magnetospirilla (SH-1, PR-1, MV-1, and QH-2) are smaller (Table 1). The G+C content of SH-1 is smaller than those of MV-1 and QH-2 but higher than that of PR-1 (Table 1). In particular, SH-1 has a higher number of tRNAs and rRNA operons (Table 1). There was no evidence for the presence of extrachromosomal elements such as plasmids for SH-1. A total of 1 847 CDS (50.8%) could be assigned to putative functions, 1 227 CDS (33.8%) represented conserved hypothetical proteins of unknown function, and the remaining 559 CDS (18.4%) show no sequence similarity to any previously reported sequence. We identified 76 tandem repeats in the genome of SH-1, including 51 minisatellite DNAs and four microsatellite DNAs. No CRISPR sequence was found. In addition, three prophages were identified to be distributed throughout the genome.
|
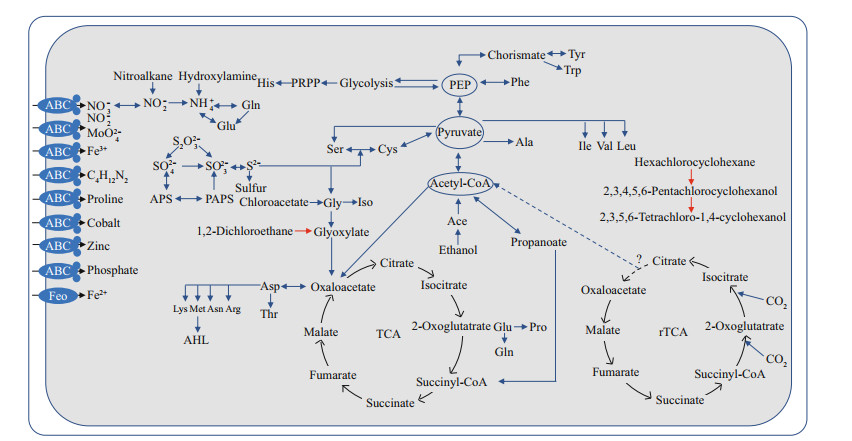
Genes involved in glycolysis (core module involving three-carbon compounds), the tricarboxylic acid (TCA) cycle, the non-oxidative phase of reductive pentose phosphate pathway, phosphoribosyl diphosphate (PRPP) biosynthesis, the ethylmalonyl pathway, and the synthesis of all 20 essential amino acids were identified in SH-1 genome (Fig. 1). Genes involved in the Embden-Meyerhof pathway in glycolysis including hexokinase (enzyme commission (EC): 2.7.1.1), glucokinase (EC: 2.7.1.2), and glyceraldehyde-3-phosphate dehydrogenase (EC: 1.2.7.6 and EC: 1.2.1.9) were missing. Most of the genes involved in reductive tricarboxylic acid (rTCA) cycle were detected, except for ATP-citrate lyase (EC: 2.3.3.8), suggesting that SH-1 is not able to use the rTCA cycle for autotrophy.
|
| Fig.1 Reconstruction of metabolic pathways in SH-1 The reconstructed glycolysis, TCA cycle, amino acid biosynthesis, nitrogen metabolism, and sulfur metabolism pathways of SH-1 are shown. Most of genes involved in the reductive tricarboxylic acid cycle (rTCA) were also present, except for ATP citrate lyase. The presence of haloalkane dehalogenase suggests the potential for SH-1 to catalyze the degradation of chlorocyclohexane, chlorobenzene, and chloroalkane (red). |
Protein encoded by gene SH1_v1_2542 was found to be 67.1% identical to a fragment (170 aa/310 aa) of the gene dhlA encoding haloalkane dehalogenase (EC: 3.8.1.5), which was previously identified and characterized in Xanthobacter autotrophicus involved in the degradation of chlorocyclohexane, chlorobenzene, chloroalkane, and chloroalkene (Fig. 1). Organisms having haloalkane dehalogenase are able to degrade hexachlorocyclohexane to 2, 3, 4, 5, 6-pentachlorocyclohexanol and 2, 3, 5, 6-tetrachloro-1, 4-cyclohexanol, and to degrade 1.2-dichloroethane to glyoxylate (Janssen et al., 1989). Similar genes have not been detected in the genomes of other MTB or other organisms belonging to genus Terasakiella. Its presence in SH-1 could be a result of horizontal gene transfer (HGT). It should be noted that active-site residues His289 and Asp260 of DHLA are not present in SH1_v1_2542, thus its function awaits further detailed characterization (Supplementary Fig.S1) (Verschueren et al., 1993). Furthermore, additional studies are needed to assess whether SH-1 has the ability to degrade chlorocyclohexane, chlorobenzene, chloroalkane, and chloroalkene, and to investigate the broader complexity of carbon metabolism by SH-1.
3.3 Sulfur metabolismUnlike AMB-1, MSR-1, and QH-2, no evidence was found in the genome of SH-1 for the presence of any of the genes for dissimilatory sulfate reduction, including dsr genes or the Apr system. However, cysN and cysD were present in the genome of SH-1, which encode enzymes that activate sulfate and catalyze the synthesis of adenosine-5 -phosphosulfate (APS) in the assimilatory sulfate reduction pathway (Fig. 1). Genes of cysC (APS kinase; produces 3 -phosphoadenosine-5 -phosphosulfate: PAPS), cysH (PAPS reductase; catalyzes the conversion of PAPS to sulfite), and cysI (sulfite reductase) were also identified. Therefore, SH-1 may be capable of assimilatory sulfate reduction to H2S (Pinto et al., 2004). SH-1 also contained soxBAZYX gene cluster involved in thiosulfate oxidation, and the soxCD genes encoding sulfur dehydrogenase were also found (Fig. 1). These findings suggest that SH-1 could use thiosulfate as an electron donor and oxidize it to sulfate (Hensen et al., 2006).
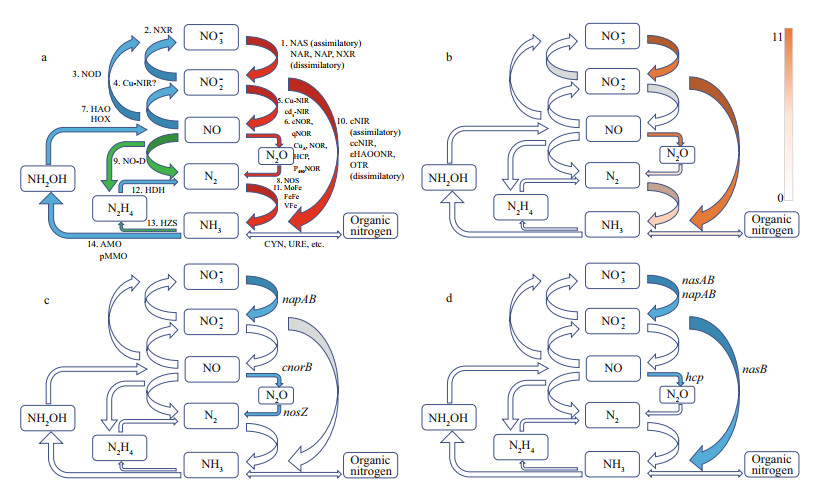
3.4 Nitrogen metabolismBoth SH-1 and PR-1 contained genes encoding periplasmic dissimilatory nitrate reductases (napAB), assimilatory nitrate reductase (nasAB), hydroxylamine reductase (hcp), and assimilatory nitrite reductases (nasB) (Fig. 1), suggesting that they can gain energy by converting nitrate to ammonia (Kuypers et al., 2018). nirB is identified in genomes of SH-1 and PR-1, but nirD is not present. Whether NirB alone could carry out the function needs more studies. napAB are present in the genomes of Terasakiella pusilla but nasAB or hcp are not identified (Fig. 2c & d). However, T. pusilla contains genes encoding cytochrome c-dependent nitric oxide (NO) reductases (cNOR and cnorB), and nitrous oxide reductases (nosZ) (Fig. 2c), which are not present in SH-1 and PR-1. This may reflect differences in the metabolism of MTB compared with other bacteria belonging to the same genus. Although genes fixS, fixG, and fixI were identified, the nif gene cluster responsible for nitrogen fixation was not present in the genomes of genus Terasakiella. Whether the genus Terasakiella can fix nitrogen needs further investigation.
|
| Fig.2 Microbial transformations of nitrogen compounds (modified from Kuypers et al., 2018) a. microbial enzymes involved in 15 reactions for eight key inorganic nitrogen species having diff erent oxidation states. Red arrow refers to reduction reaction, blue arrow refers to oxidation reaction, and green arrow refers to disproportionation or comproportionation reaction; b. the reactions found in MTB. The heatmap shows the rate of reactions; c. the reactions found in Terasakiella pusilla (blue); d. the reactions found in SH - 1 (blue). |
We further compared the microbial transformations of nitrogenamong14reprentativespecies of MTB (Terasakiella sp.SH-1, Candidatus Terasakiella magnetica strainPR-1, Magnetospirasp.strainQH-2, Magnetospirillum gryphiswaldense strainMSR-1, Magnetospirillum magneticum strainAMB-1, Magnetococcus marinus MC-1, Magneto-ovoid bacterium MO-1, Ectothiorhodospiraceae bacterium BW -2, Gammaproteobacteria magnetotactic strain SS -5, Desulfovibrio magneticus RS-1, Candidatus Magnetomorum sp.HK-1, Candidatus Magnetoglobus multicellularis str. Araruama, Candidatus Magnetobacterium bavaricumand Candidatus Magnetoovum chiemensis)(Fig. 2a & b).Seven reactions have been found in one or more genome sof the tested microorganisms(Fig. 2b), including nitrate reductase (nasA, narGH, and napA), heme-containing nitrite reductases(nirS), nitric oxide reductase(hcp, cnorB, and norVW), nitrous oxide reductase(nosZ), assimilatory (nasB and nirBD) anddissimilatory nitrite reductase(nrfAHandOTR), nitrogenases (nifHDK), cyanase(cynS), andurease(ureABC).
Among these enzymes, genes encoding for nitrate reductases, nitrite reductase and nitric oxide reductases were most commonly found.These genes occur widely in genomes of Alphaproteobacteria, Gammaproteobacteria, and Etaproteobacteria. Our results show that MTB commonly act as denitrifiers and nitrogen-fixers in nitrogen cycle processes. Previous studies have shown the potential link between denitrification and magnetosome formation in freshwater Magnetospirillum spp. (Bazylinski and Blakemore, 1983; Matsunaga et al., 1991; Matsunaga and Tsujimura, 1993; Yang et al., 2001; Li et al., 2012). Therefore, denitrification should play an important role in redox control for magnetosome formation. However, their role in magnetosome formation has not yet been fully clarified and needs further experimental investigation.
3.5 ProphageTwo predicted prophage regions were found in the genome of SH-1, having lengths of 11.0 22.6 kb and G+C contents of 46.1% 48.4% (Supplementary Table S2 & Fig.S2). Among these, SH1_ v1_1640 1665 comprised 23 CDS containing three predicted phage tail collar domain proteins, two plasmid maintenance system killer proteins (HigB), and seven transposases, one sulfotransferase and seven hypothetical proteins (Supplementary Fig.S2a & b1), while SH1_v1_2382 to SH1_v1_2398 comprised 16 CDS, including three putative tail-related proteins, one putative endolysin, one DNA maturase beta subunit, one putative major capsid protein, and one putative GcrA-like cell cycle regulator (Supplementary Fig.S2a & 2b). No genes encoding lysine and holin, representing the lysis module, were found.
We searched genomes of 12 bacteria (Terasakiella sp. SH-1, Candidatus Terasakiella magnetica strain PR-1, Magnetospira sp. strain QH-2, Magnetospirillum gryphiswaldense strain MSR-1, Magnetospirillum magneticum strain AMB-1, Magnetococcus marinus MC-1, Magneto-ovoid bacterium MO-1, Desulfovibrio magneticus RS-1, Candidatus Magnetomorum sp. HK-1, Candidatus Magnetoglobus multicellularis str. Araruama, Candidatus Magnetobacterium bavaricum, and Candidatus Desulfamplus magnetomortis BW-1) using PHASTER (Supplementary Table S3). Most MTB genomes (11 out of 12 genomes) were found to contain predicted prophage regions, with HK-1 being the only exception. Temperate phage genes have been identified in 40%–50% of microbial genomes (Canchaya et al., 2003; Casjens, 2003; Fouts, 2006; Paul, 2008; Touchon et al., 2016), and almost 50% of bacterial genomes contain at least one prophage (Touchon et al., 2016). Our results show that prophages may exist in many genomes of MTB. Through lysogenic conversion or transduction, bacteriophages and archaeal viruses contribute to the horizontal transfer of genetic material among microbial genomes (Touchon et al., 2016, 2017). Previous studies have shown that HGT plays an important role in the evolution of magnetotaxis in bacteria (Rioux et al., 2010; Monteil et al., 2018). Alternatively, being infected by phage may contribute to this process.
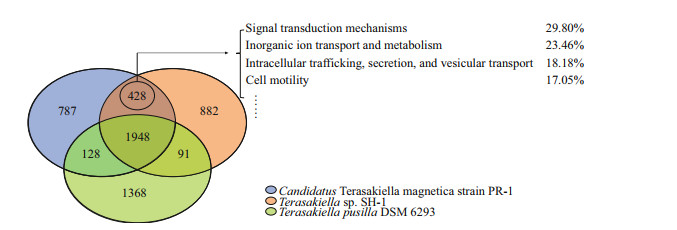
3.6 Comparative gene content analysis of three Terasakiella strains using reciprocal best matchesWe further perform pangenome analysis of SH-1, PR-1, and T. pusilla. As shown in Fig. 3, 1948 gene families were shared among the three species, representing approximately 50% of the proteins in each strain. A total of 428 families were shared by SH-1 and PR-1, but not T. pusilla. The larger overlap between SH-1 and PR-1 suggests that they are more closely related to each other than to T.pusilla. These are mainly involved in signal transduction mechanisms (29.80%), inorganic ion transport and metabolism(23.46%), intracellular trafficking, secretion and vesicular transport (18.18%) and cell motility (17.05%) (according to COG automatic classification of Microscope). These genes may also be associated with magnetosome biosynthesis.
|
| Fig.3 Venn diagram of pan/core gene analysis in the genus Terasakiella PR-1 (blue), Terasakiella pusilla (green), and SH-1(orange). The numbers indicate the number of gene families. |
Interestingly, genes related to iron metabolism have been found in the genome of T. pusilla but not in SH-1 and PR-1 genomes. These genes included fecR (Anti-FecI sigma factor), fbpC (Fe3+ ion import ATP-binding protein) and bfr (bacterioferritin, iron storage and detoxification protein). Furthermore, we found 41 copies of histidine kinase in the genome of T. pusilla, fewer than that in SH-1 (74) and PR-1 (70). However, more copies of methyl-accepting chemotaxis proteins (MCPs) were found in T. pusilla (69) than in SH-1(59) and PR-1 (57), consistent with previous report that high numbers of MCPs are not a common feature of MTB (Ji et al., 2014).
4 CONCLUSIONComparative genomic analysis of SH-1 performed in the present study revealed that genes involved in glycolysis, the TCA cycle, the non-oxidative phase of the reductive pentose phosphate pathway, PRPP biosynthesis, the ethylmalonyl pathway, and the synthesis of all 20 essential amino acids were identified. A fragment of gene encoding haloalkane dehalogenase was also identified. The soxBAZYX gene cluster, soxCD genes, and cysCHI genes were present, indicating that SH-1 is capable of assimilatory sulfate reduction to H2S, and using thiosulfate as electron donor and oxidizing it to sulfate. SH-1 also contained napAB, nasA and nasB, suggesting that it can gain energy by converting nitrate to ammonia. Two predicted prophage regions were found in SH-1, and our results suggest that prophage may be common in MTB. Pangenome analysis showed that differences between T. pusilla and both SH-1 and PR-1 may be related to signal transduction mechanisms, inorganic ion transport and metabolism, intracellular trafficking, secretion, vesicular transport and cell motility.
5 DATA AVAILABILITY STATEMENTAll data generated and analyzed during the current study are available from the corresponding author upon request.
6 ACKNOWLEDGMENTWe thank Jianhong XU from Institute of Oceanology, Chinese Academy of Sciences and Weijia ZHANG from Institute of Deep-sea Science and Engineering, Chinese Academy of Sciences, for assistance in sampling.
Electronic supplementary material Supplementary material (Supplementary Tables S1–S3 and Figs.S1–S2) is available in the online version of this article at https://doi.org/10.1007/s00343-021-1054-5.
Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. 1990. Basic local alignment search tool. Journal of Molecular Biology, 215(3): 403-410.
DOI:10.1016/S0022-2836(05)80360-2 |
Apweiler R, Bairoch A, Wu C H, Barker W C, Boeckmann B, Ferro S, Gasteiger E, Huang H Z, Lopez R, Magrane M, Martin M J, Natale D A, O'Donovan C, Redaschi N, Yeh L S L. 2004. UniProt: the Universal Protein knowledgebase. Nucleic Acids Research, 32(S1): D115-D119.
DOI:10.1093/nar/gkh131 |
Arndt D, Grant J R, Marcu A, Sajed T, Pon A, Liang Y J, Wishart D S. 2016. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Research, 44(W1): W16-W21.
DOI:10.1093/nar/gkw387 |
Ashburner M, Ball C A, Blake J A, Botstein D, Butler H, Cherry J M, Davis A P, Dolinski K, Dwight S S, Eppig J T, Harris M A, Hill D P, Tarver L I, Kasarskis A, Lewis S, Matese J C, Richardson J E, Ringwald M, Rubin G M, Sherlock G. 2000. Gene Ontology: tool for the unification of biology. Nature Genetics, 25(1): 25-29.
DOI:10.1038/75556 |
Bazylinski D A, Blakemore R P. 1983. Denitrification and assimilatory nitrate reduction in Aquaspirillum magnetotacticum. Applied and Environmental Microbiology, 46(5): 1118-1124.
DOI:10.1128/aem.46.5.1118-1124.1983 |
Bazylinski D A, Frankel R B, Heywood B R, Mann S, King J W, Donaghay P L, Hanson A K. 1995. Controlled biomineralization of magnetite (Fe3O4) and greigite(Fe3S4) in a magnetotactic bacterium. Applied and Environmental Microbiology, 61: 3232-3239.
DOI:10.1128/aem.61.9.3232-3239.1995 |
Bazylinski D A, Frankel R B. 2004. Magnetosome formation in prokaryotes. Nature Reviews Microbiology, 2(3): 217-230.
DOI:10.1038/nrmicro842 |
Bazylinski D A, Williams T J, Lefèvre C T, Trubitsyn D, Fang J S, Beveridge T J, Moskowitz B M, Ward B, Schübbe S, Dubbels B L, Simpson B. 2013. Magnetovibrio blakemorei gen.nov., sp. nov., a magnetotactic bacterium(Alphaproteobacteria: Rhodospirillaceae) isolated from a salt marsh. International Journal of Systematic and Evolutionary Microbiology, 63(Pt 5): 1824-1833.
DOI:10.1099/ijs.0.044453-0 |
Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Research, 27(2): 573-580.
DOI:10.1093/nar/27.2.573 |
Blakemore R P, Maratea D, Wolfe R S. 1979. Isolation and pure culture of a freshwater magnetic spirillum in chemically defined medium. Journal of Bacteriology, 140(2): 720-729.
DOI:10.1128/jb.140.2.720-729.1979 |
Blakemore R P. 1982. Magnetotactic bacteria. Annual Review of Microbiology, 36: 217-238.
DOI:10.1146/annurev.mi.36.100182.001245 |
Canchaya C, Proux C, Fournous G, Bruttin A, Brüssow H. 2003. Prophage genomics. Microbiology and Molecular Biology Reviews, 67(2): 238-276.
DOI:10.1128/MMBR.67.2.238-276.2003 |
Casjens S. 2003. Prophages and bacterial genomics: what have we learned so far?. Molecular Microbiology, 49(2): 277-300.
DOI:10.1046/j.1365-2958.2003.03580.x |
Consortium U P. 2015. UniProt: a hub for protein information. Nucleic Acids Research, 43(D1): D204-D212.
DOI:10.1093/nar/gku989 |
DeLong E F, Frankel R B, Bazylinski D A. 1993. Multiple evolutionary origins of magnetotaxis in bacteria. Science, 259(5096): 803-806.
DOI:10.1126/science.259.5096.803 |
Du H J, Zhang W Y, Zhang W S, Zhang W J, Pan H M, Pan Y X, Bazylinski D A, Wu L F, Xiao T, Lin W. 2019. Magnetosome gene duplication as an important driver in the evolution of magnetotaxis in the Alphaproteobacteria. mSystems, 4(5): e00315-19.
DOI:10.1128/mSystems.00315-19 |
Fouts D E. 2006. Phage_Finder: automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Research, 34(20): 5839-5851.
DOI:10.1093/nar/gkl732 |
Galperin M Y, Makarova K S, Wolf Y I, Koonin E V. 2015. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Research, 43(D1): D261-D269.
DOI:10.1093/nar/gku1223 |
Geurink C, Lefèvre C T, Monteil C L, Morillo-Lopez V, Abreu F, Bazylinski D A, Trubitsyn D. 2020. Complete genome sequence of strain BW-2, a magnetotactic gammaproteobacterium in the family Ectothiorhodospiraceae, isolated from a brackish spring in Death Valley, California. Microbiology Resource Announcements, 9(1): e01144-19.
DOI:10.1128/MRA.01144-19 |
Grissa I, Vergnaud G, Pourcel C. 2007. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Research, 35(S2): W52-W57.
DOI:10.1093/nar/gkm360 |
Grünberg K, Wawer C, Tebo B M, Schüler D. 2001. A large gene cluster encoding several magnetosome proteins is conserved in different species of magnetotactic bacteria. Applied and Environmental Microbiology, 67(10): 4573-4582.
DOI:10.1128/AEM.67.10.4573-4582.2001 |
Hanzlik M, Winklhofer M, Petersen N. 2002. Pulsed-fieldremanence measurements on individual magnetotactic bacteria. Journal of Magnetism and Magnetic Materials, 248(2): 258-267.
DOI:10.1016/S0304-8853(02)00353-0 |
Hensen D, Sperling D, Trüper H G, Brune D C, Dahl C. 2006. Thiosulphate oxidation in the phototrophic sulphur bacterium Allochromatium vinosum. Molecular Microbiology, 62(3): 794-810.
DOI:10.1111/j.1365-2958.2006.05408.x |
Huerta-Cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter M C, Rattei T, Mende D R, Sunagawa S, Kuhn M, Jensen L J, von Mering C, Bork P. 2016. eggNOG 4.5:a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Research, 44(D1): D286-D293.
DOI:10.1093/nar/gkv1248 |
Janssen D B, Pries F, van der Ploeg J, Kazemier B, Terpstra P, Witholt B. 1989. Cloning of 1, 2-dichloroethane degradation genes of Xanthobacter autotrophicus GJ10 and expression and sequencing of the dhlA gene. Journal of Bacteriology, 171(12): 6791-6799.
DOI:10.1128/jb.171.12.6791-6799.1989 |
Ji B Y, Zhang S D, Arnoux P, Rouy Z, Alberto F, Philippe N, Murat D, Zhang W J, Rioux J B, Ginet N, Sabaty M, Mangenot S, Pradel N, Tian J S, Yang J, Zhang L C, Zhang W Y, Pan H M, Henrissat B, Coutinho P M, Li Y, Xiao T, Médigue C, Barbe V, Pignol D, Talla E, Wu L F. 2014. Comparative genomic analysis provides insights into the evolution and niche adaptation of marine Magnetospira sp.QH-2 strain. Environmental Microbiology, 16(2): 525-544.
DOI:10.1111/1462-2920.12180 |
Ji B Y, Zhang S D, Zhang W J, Rouy Z, Alberto F, Santini C L, Mangenot S, Gagnot S, Philippe N, Pradel N, Zhang L C, Tempel S, Li Y, Médigue C, Henrissat B, Coutinho P M, Barbe V, Talla E, Wu L F. 2017. The chimeric nature of the genomes of marine magnetotactic coccoid-ovoid bacteria defines a novel group of Proteobacteria. Environmental Microbiology, 19(3): 1103-1119.
DOI:10.1111/1462-2920.13637 |
Jogler C, Schüler D. 2009. Genomics, genetics, and cell biology of magnetosome formation. Annual Review of Microbiology, 63: 501-521.
DOI:10.1146/annurev.micro.62.081307.162908 |
Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Research, 44(D1): D457-D462.
DOI:10.1093/nar/gkv1070 |
Kolinko S, Jogler C, Katzmann E, Wanner G, Peplies J, Schüler D. 2012. Single-cell analysis reveals a novel uncultivated magnetotactic bacterium within the candidate division OP3. Environmental Microbiology, 14(7): 1709-1721.
DOI:10.1111/j.1462-2920.2011.02609.x |
Kuypers M M M, Marchant H K, Kartal B. 2018. The microbial nitrogen-cycling network. Nature Reviews Microbiology, 16(5): 263-276.
DOI:10.1038/nrmicro.2018.9 |
Lefèvre C T, Abreu F, Schmidt M L, Lins U, Frankel R B, Hedlund B P, Bazylinski D A. 2010. Moderately thermophilic magnetotactic bacteria from hot springs in Nevada. Applied and Environmental Microbiology, 76(11): 3740-3743.
DOI:10.1128/AEM.03018-09 |
Lefèvre C T, Bazylinski D A. 2013. Ecology, diversity, and evolution of magnetotactic bacteria. Microbiology and Molecular Biology Reviews, 77(3): 497-526.
DOI:10.1128/MMBR.00021-13 |
Lefèvre C T, Bernadac A, Yu-Zhang K, Pradel N, Wu L F. 2009. Isolation and characterization of a magnetotactic bacterial culture from the Mediterranean Sea. Environmental Microbiology, 11(7): 1646-1657.
DOI:10.1111/j.1462-2920.2009.01887.x |
Li Y J, Katzmann E, Borg S, Schüler D. 2012. The periplasmic nitrate reductase nap is required for anaerobic growth and involved in redox control of magnetite biomineralization in Magnetospirillum gryphiswaldense. Journal of Bacteriology, 194(18): 4847-4856.
DOI:10.1128/JB.00903-12 |
Lin W, Pan Y X, Bazylinski D A. 2017a. Diversity and ecology of and biomineralization by magnetotactic bacteria. Environmental Microbiology Reports, 9(4): 345-356.
DOI:10.1111/1758-2229.12550 |
Lin W, Pan Y X. 2015. A putative greigite-type magnetosome gene cluster from the candidate phylum Latescibacteria. Environmental Microbiology Reports, 7(2): 237-242.
DOI:10.1111/1758-2229.12234 |
Lin W, Paterson G A, Zhu Q Y, Wang Y Z, Kopylova E, Li Y, Knight R, Bazylinski D A, Zhu R X, Kirschvink J L, Pan Y X. 2017b. Origin of microbial biomineralization and magnetotaxis during the Archean. Proceedings of the National Academy of Sciences of the United States of America, 114(9): 2171-2176.
DOI:10.1073/pnas.1614654114 |
Lin W, Zhang W S, Paterson G A, Zhu Q Y, Zhao X, Knight R, Bazylinski D A, Roberts A P, Pan Y X. 2020. Expanding magnetic organelle biogenesis in the domain Bacteria. Microbiome, 8(1): 152.
DOI:10.1186/s40168-020-00931-9 |
Lin W, Zhang W S, Zhao X, Roberts A P, Paterson G A, Bazylinski D A, Pan Y X. 2018. Genomic expansion of magnetotactic bacteria reveals an early common origin of magnetotaxis with lineage-specific evolution. The ISME Journal, 12(6): 1508-1519.
DOI:10.1038/s41396-018-0098-9 |
Lohße A, Ullrich S, Katzmann E, Borg S, Wanner G, Richter M, Voigt B, Schweder T, Schüler D. 2011. Functional analysis of the magnetosome island in Magnetospirillum gryphiswaldense: the mamAB operon is sufficient for magnetite biomineralization. PLoS One, 6(10): e25561.
DOI:10.1371/journal.pone.0025561 |
Makarova K S, Wolf Y I, Koonin E V. 2015. Archaeal clusters of orthologous genes (arcogs): an update and application for analysis of shared features between thermococcales, methanococcales, and methanobacteriales. Life, 5(1): 818-840.
DOI:10.3390/life5010818 |
Maratea D, Blakemore R P. 1981. Aquaspirillum magnetotacticum sp.nov., a magnetic spirillum. International Journal of Systematic and Evolutionary Microbiology, 31(4): 452-455.
DOI:10.1099/00207713-31-4-452 |
Matsunaga T, Okamura Y, Fukuda Y, Wahyudi A T, Murase Y, Takeyama H. 2005. Complete genome sequence of the facultative anaerobic magnetotactic bacterium Magnetospirillum sp.strain AMB-1. DNA Research, 12(3): 157-166.
DOI:10.1093/dnares/dsi002 |
Matsunaga T, Sakaguchi T, Tadakoro F. 1991. Magnetite formation by a magnetic bacterium capable of growing aerobically. Applied Microbiology and Biotechnology, 35(5): 651-655.
DOI:10.1007/BF00169632 |
Matsunaga T, Tsujimura N. 1993. Respiratory inhibitors of a magnetic bacterium Magnetospirillum sp.AMB-1 capable of growing aerobically. Applied Microbiology and Biotechnology, 39(3): 368-371.
DOI:10.1007/BF00192094 |
Miele V, Penel S, Duret L. 2011. Ultra-fast sequence clustering from similarity networks with SiLiX. BMC Bioinformatics, 12: 116.
DOI:10.1186/1471-2105-12-116 |
Monteil C L, Perrière G, Menguy N, Ginet N, Alonso B, Waisbord B, Cruveiller S, Pignol D, Lefèvre C T. 2018. Genomic study of a novel magnetotactic Alphaproteobacteria uncovers the multiple ancestry of magnetotaxis. Environmental Microbiology, 20(12): 4415-4430.
DOI:10.1111/1462-2920.14364 |
Murat D, Quinlan A, Vali H, Komeili A. 2010. Comprehensive genetic dissection of the magnetosome gene island reveals the step-wise assembly of a prokaryotic organelle. Proceedings of the National Academy of Sciences of the United States of America, 107(12): 5593-5598.
DOI:10.1073/pnas.0914439107 |
Paul J H. 2008. Prophages in marine bacteria: dangerous molecular time bombs or the key to survival in the seas?. The ISME Journal, 2(6): 579-589.
DOI:10.1038/ismej.2008.35 |
Pinto R, Tang Q X, Britton W J, Leyh T S, Triccas J A. 2004. The Mycobacterium tuberculosis cysD and cysNC genes form a stress-induced operon that encodes a tri-functional sulfate-activating complex. Microbiology, 150(6): 1681-1686.
DOI:10.1099/mic.0.26894-0 |
Richter M, Kube M, Bazylinski D A, Lombardot T, Glöckner F O, Reinhardt R, Schüler D. 2007. Comparative genome analysis of four magnetotactic bacteria reveals a complex set of group-specific genes implicated in magnetosome biomineralization and function. Journal of Bacteriology, 189(13): 4899-4910.
DOI:10.1128/JB.00119-07 |
Rioux J B, Philippe N, Pereira S, Pignol D, Wu L F, Ginet N. 2010. A second actin-like MamK protein in Magnetospirillum magneticum AMB-1 encoded outside the genomic magnetosome island. PLoS One, 5(2): e9151.
DOI:10.1371/journal.pone.0009151 |
Sakaguchi T, Arakaki A, Matsunaga T. 2002. Desulfovibrio magneticus sp.nov., a novel sulfate-reducing bacterium that produces intracellular single-domain-sized magnetite particles. International Journal of Systematic and Evolutionary Microbiology, 52(Pt 1): 215-221.
DOI:10.1099/00207713-52-1-215 |
Schleifer K H, Schüler D, Spring S, Weizenegger M, Amann R, Ludwig W, Köhler M. 1991. The genus Magnetospirillum gen.nov. description of Magnetospirillum gryphiswaldense sp. nov. and transfer of Aquaspirillum magnetotacticum to Magnetospirillum magnetotacticum comb. nov. 1. Systematic and Applied Microbiology, 14(4): 379-385.
DOI:10.1016/s0723-2020(11)80313-9 |
Schübbe S, Williams T J, Xie G, Kiss H E, Brettin T S, Martinez D, Ross C A, Schüler D, Cox B L, Nealson K H, Bazylinski D A. 2009. Complete genome sequence of the chemolithoautotrophic marine magnetotactic coccus strain MC-1. Applied Environmental Microbiology, 75(14): 4835-4852.
DOI:10.1128/AEM.02874-08 |
Simmons S L, Sievert S M, Frankel R B, Bazylinski D A, Edwards K J. 2004. Spatiotemporal distribution of marine magnetotactic bacteria in a seasonally stratified coastal salt pond. Applied and Environmental Microbiology, 70(10): 6230-6239.
DOI:10.1128/AEM.70.10.6230-6239.2004 |
Touchon M, Bernheim A, Rocha E P C. 2016. Genetic and lifehistory traits associated with the distribution of prophages in bacteria. The ISME Journal, 10(11): 2744-2754.
DOI:10.1038/ismej.2016.47 |
Touchon M, de Sousa J A M, Rocha E P C. 2017. Embracing the enemy: the diversification of microbial gene repertoires by phage-mediated horizontal gene transfer. Current Opinion in Microbiology, 38: 66-73.
DOI:10.1016/j.mib.2017.04.010 |
Trubitsyn D, Monteil C L, Geurink C, Morillo-Lopez V, de Almeida L G P, de Vasconcelos T R, Abreu F, Bazylinski D A, Lefevre C T. 2021. Complete genome sequence of strain SS-5, a magnetotactic gammaproteobacterium isolated from the Salton Sea, a Shallow, Saline, Endorheic Rift Lake located on the San Andreas Fault in California. Microbiology Resource Announcements, 10(1): e00928-20.
DOI:10.1128/MRA.00928-20 |
Uebe R, Schüler D, Jogler C, Wiegand S. 2018. Reevaluation of the complete genome sequence of Magnetospirillum gryphiswaldense MSR-1 with single-molecule real-time sequencing data. Genome Announcements, 6(17): e00309-18.
DOI:10.1128/genomeA.00309-18 |
Uzun M, Alekseeva L, Krutkina M, Koziaeva V, Grouzdev D. 2020. Unravelling the diversity of magnetotactic bacteria through analysis of open genomic databases. Scientific Data, 7(1): 252.
DOI:10.1038/s41597-020-00593-0 |
Vali H, Förster O, Amarantidis G, Petersen N. 1987. Magnetotactic bacteria and their magnetofossils in sediments. Earth and Planetary Science Letters, 86(2-4): 389-400.
DOI:10.1016/0012-821X(87)90235-4 |
Vallenet D, Belda E, Calteau A, Cruveiller S, Engelen S, Lajus A, Le Fèvre F, Longin C, Mornico D, Roche D, Rouy Z, Salvignol G, Scarpelli S, Smith A A T, Weiman M, Médigue C. 2013. MicroScope-an integrated microbial resource for the curation and comparative analysis of genomic and metabolic data. Nucleic Acids Research, 41(D1): D636-D647.
DOI:10.1093/nar/gks1194 |
Verschueren K H G, Franken S M, Rozeboom H J, Kalk K H, Dijkstra B W. 1993. Refined X-ray structures of haloalkane dehalogenase at pH 6.2 and pH 8.2 and implications for the reaction mechanism . Journal of Molecular Biology, 232(3): 856-872.
DOI:10.1006/jmbi.1993.1436 |
Wang Y Z, Zhang T W, Lin W, Zhang B F, Cai Y, Yang C Y, Li J H, Xu H T, Pan Y X. 2016. Complete genome sequence of Magnetospirillum sp.strain XM-1, isolated from the Xi'an City moat, China. Genome Announcements, 4(5): e01171-16.
DOI:10.1128/genomeA.01171-16 |
Wenter R, Wanner G, Schüler D, Overmann J. 2009. Ultrastructure, tactic behaviour and potential for sulfate reduction of a novel multicellular magnetotactic prokaryote from North Sea sediments. Environmental Microbiology, 11(6): 1493-1505.
DOI:10.1111/j.1462-2920.2009.01877.x |
Williams T J, Lefèvre C T, Zhao W D, Beveridge T J, Bazylinski D A. 2012. Magnetospira thiophila gen. nov., sp. nov., a marine magnetotactic bacterium that represents a novel lineage within the Rhodospirillaceae(Alphaproteobacteria). International Journal of Systematic and Evolutionary Microbiology, 62(Pt 10): 2443-2450.
DOI:10.1099/ijs.0.037697-0 |
Yang C D, Takeyama H, Tanaka T, Matsunaga T. 2001. Effects of growth medium composition, iron sources and atmospheric oxygen concentrations on production of luciferase-bacterial magnetic particle complex by a recombinant Magnetospirillum magneticum AMB-1. Enzyme and Microbial Technology, 29(1): 13-19.
DOI:10.1016/S0141-0229(01)00343-X |
Zhou K, Zhang W Y, Pan H M, Li J H, Yue H D, Xiao T, Wu L F. 2013. Adaptation of spherical multicellular magnetotactic prokaryotes to the geochemically variable habitat of an intertidal zone. Environmental Microbiology, 15(5): 1595-1605.
DOI:10.1111/1462-2920.12057 |
Zhu K L, Pan H M, Li J H, Yu-Zhang K, Zhang S D, Zhang W Y, Zhou K, Yue H D, Pan Y X, Xiao T, Wu L F. 2010. Isolation and characterization of a marine magnetotactic spirillum axenic culture QH-2 from an intertidal zone of the China Sea. Research in Microbiology, 161(4): 276-283.
DOI:10.1016/j.resmic.2010.02.003 |
 2021, Vol. 39
2021, Vol. 39


