Institute of Oceanology, Chinese Academy of Sciences
Article Information
- GONG Lin, LIM Swee-Cheng, YANG Mei, LI Xinzheng
- A new species of Macandrewia (Demospongiae, Tetractinellida, Macandrewiidae) from a seamount in the Western Pacific Ocean
- Journal of Oceanology and Limnology, 39(5): 1730-1739
- http://dx.doi.org/10.1007/s00343-021-1042-9
Article History
- Received Feb. 4, 2021
- accepted in principle Feb. 25, 2021
- accepted for publication May. 27, 2021
2 Center for Ocean Mega-Science, Chinese Academy of Sciences, Qingdao 266071, China;
3 Laboratory for Marine Biology and Biotechnology, Pilot National Laboratory for Marine Science and Technology (Qingdao), Qingdao 266237, China;
4 University of Chinese Academy of Sciences, Beijing 100049, China;
5 National University of Singapore, Tropical Marine Science Institute, Singapore 119227, Singapore
The family Macandrewiidae Schrammen, 1924 has a rigid skeleton formed by desmas that confers a very strong skeleton and hard texture. It is one of the 13 families of 'Lithistid' demosponges (order Tetractinellida Marshall, 1876) scattered across suborders Astrophorina Sollas, 1887 and Spirophorina Bergquist & Hogg, 1969 based on molecular data and the clear morphological synapomorphies are lacking (Morrow and Cárdenas, 2015).
At present, Macandrewiidae contains only one genus, Macandrewia Gray, 1859, and nine valid species (van Soest et al., 2021) characterized by the presence of dentate phyllotriaenes/discotriaenes, smooth oxeas as ectosomal megascleres, desmas with a triaenose crepis and microxeas as microscleres (Pisera and Lévi, 2002)). Majority of Macandrewia species have been described from the Atlantic Ocean (M. azorica Gray, 1859; M. clavatella (Schmidt, 1870); M. ramosa Topsent, 1904; M. robusta Topsent, 1904; M. minima Carvalho & Xavier, 2020; and M. schusterae Carvalho & Xavier, 2020). Only two species (M. spinifoliata Lévi & Lévi, 1983; M. rigida Lévi & Lévi, 1989) have been reported from the Pacific Ocean and one species (M. auris Lendenfeld, 1907) from the Indian Ocean.
A new species of Macandrewia was discovered during a biodiversity survey cruise of seamounts near Yap Trench (Western Pacific Ocean) in December 2014 conducted by the Institute of Oceanology, Chinese Academy of Sciences (IOCAS), using the R/V Kexue (Science in Chinese). The new sponge species was collected between the depth of 255- 311 m. Morphological examination and molecular analysis of the specimen revealed that it is a new species of Macandrewia, the third recorded for the Pacific Ocean.
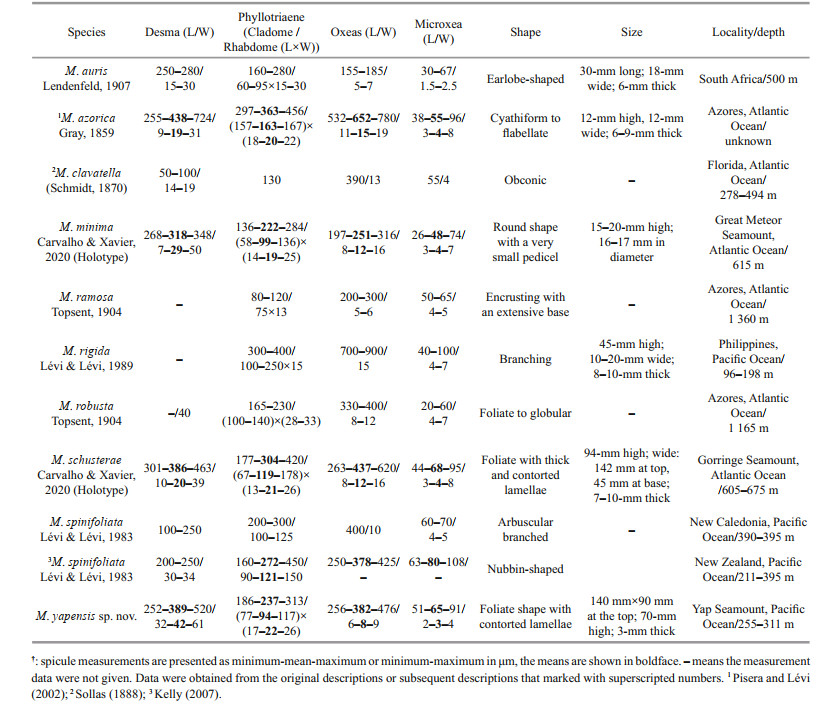
2 MATERIAL AND METHOD 2.1 Sample collectionThe sponge specimen was collected by the remotely operated vehicle (ROV) Faxian (Discovery in Chinese) from a seamount near Yap Trench in the Western Pacific Ocean, and upon collection it was preserved in 75% ethanol and described. The sample was deposited in the Marine Biological Museum of Chinese Academy of Sciences (MBMCAS), Qingdao, China, using accession prefix MBM286832. Ocean Data View software version 5.4.0 (Schlitzer, 2020) was used to draw the collection locations (Fig. 1).
|
| Fig.1 The locations of Macandrewia species found in Pacific Ocean Based on the data of references (Lévi and Lévi, 1983, 1989). |
Spicules were isolated by digesting a small piece of sponge tissue using concentrated nitric acid at room temperature overnight. Spicules and skeletal structures were coated with gold and observed under a Hitachi S-3400N scanning electron microscope (SEM). Spicules were measured using an Olympus DSX500 optodigital light microscope (LM). Measurement results are given in Table 1.

|
Total genomic DNA was extracted from sponge tissue using Tissue DNA Kit (OMEGA Qingdao, China). Polymerase chain reaction (PCR) amplification was carried out in a 25-μL total reaction volume containing 12.5 μL of Premix TaqTM (TaKaRa, Japan), 1 μL of each primer, 2 μL of template DNA, and 8.5 μL of DNase-free ddH2O. The COI segment was amplified with the degenerated Folmer primers (dgLCO1490 and dgHCO2198) designed by Meyer et al. (2005). Amplification was performed using the following procedure: initial denaturation for 5 min at 94 ℃, followed by 30 cycles of denaturation for 30 s at 94 ℃, annealing for 40 s at 48 ℃, extension for 30 s at 72 ℃, and a final extension for 5 min at 72 ℃. The COI sequence was submitted to GenBank with an accession number of MW543306.
2.4 Phylogenetic analysisSpecimen was analyzed based on the partial COI gene sequences, by which the phylogenetic position of Macandrewia yapensis sp. nov. in relation with other members of Astrophorina was clarified. There are 64 genera and 15 families in Astrophorina (van Soest et al., 2021) but only 39 genera in 14 families are available from GenBank. We downloaded one sequence from each of the 39 genera to build the phylogenetic tree (Table 2) including the sequences of Macandrewia azorica from the Azores archipelago and two undescribed Macandrewia sp. from South Africa, and Scleritoderma flabelliforme was used as the outgroup for the construction of the phylogenetic tree.

|
The homologous sequences including 43 sequences of COI genes were aligned using MUSCLE 3.8 (Edgar, 2004) with default parameters. The best fitting nucleotide substitution model was GTR+I+G as determined by ModelTest 3.7 (Posada and Crandall, 1998) with the AIC (Akaike Information Criterion). Maximum likelihood (ML) analysis was carried out in PhyloSuite (Zhang et al., 2020) batch mode using IQ-TREE (Trifinopoulos et al., 2016) plug-in in "Auto" mode with 200 000 ultrafast bootstrap replicates. Bayesian inference (BI) analysis was conducted using Mrbayes 3.2 (Huelsenbeck and Ronquist, 2001); Markov Chains were run for 10 million generations, with sampling every 1 000 generations. The first 25% of trees were discarded as burn-in, and the remaining trees were summarized in 50% majority rule consensus tree to estimate the posterior probabilities. Tracer v1.7 (Rambaut et al., 2018) was used to determine the effective sample size (ESS) values for all sampled parameters to ensure that convergence was reached. The trees were edited using iTOL (https://itol.embl.de/) (Letunic and Bork, 2019) and the branch lengths were ignored for presentation purpose.
3 TAXONOMYClass Demospongiae Sollas, 1885 Order Tetractinellida Marshall, 1876 Suborder Astrophorina Sollas, 1887 Family Macandrewiidae Schrammen, 1924 Genus Macandrewia Gray, 1859 Macandrewia yapensis sp. nov. (Figs. 2-4; Table 1)

|
| Fig.2 Macandrewia yapensis sp. nov (holotype MBM286832) a. the in-situ growth form of the specimen on the seabed; b. external morphology of the specimen after being removed out of seawater. |
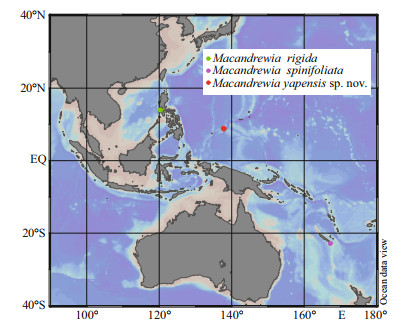
|
| Fig.3 Macandrewia yapensis sp. nov., holotype MBM286832, the skeleton a-b. overview of the outer ectosomal surface showing oxeas and phyllotriaenes; c. overview of choanosomal skeleton; d. choanosomal desmas with short, rounded, and blunt tubercles; e. details of desmas; f. desma zygosis. |
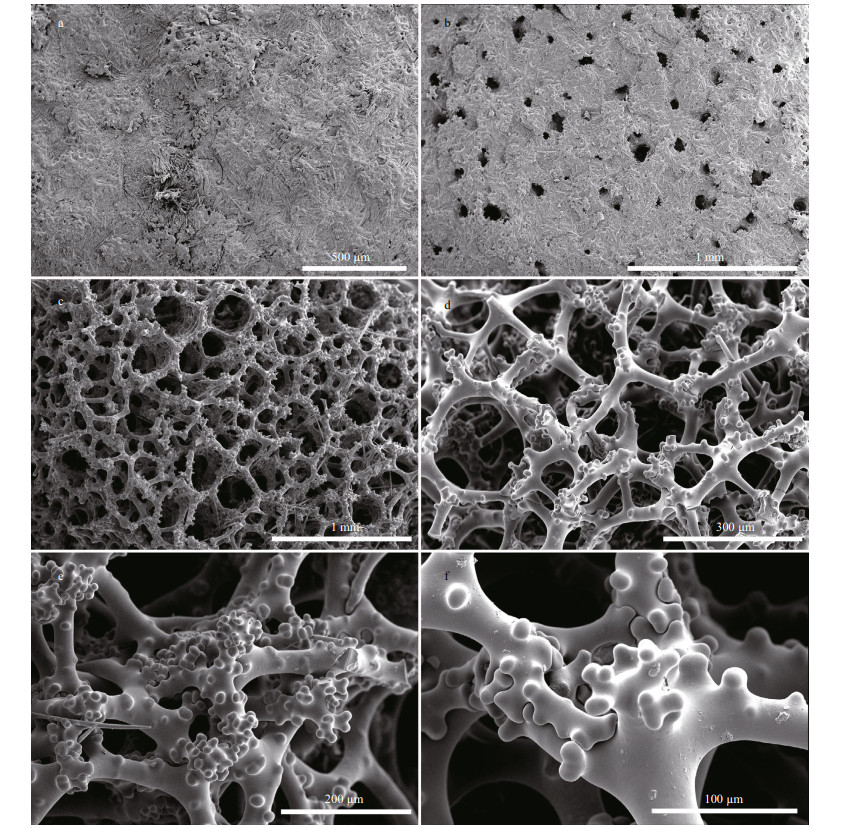
|
| Fig.4 Macandrewia yapensis sp.nov., holotype MBM286832, spicule complement a. oxea; b-c. phyllotriaenes; d. desma; e. microxea. |
Holotype: MBM286832, seamount near Yap Trench (8°51'N, 137°47'E), Western Pacific Ocean, 22 December 2014, 255-311 m, hard bottom with a thin layer of sediment and sand. The entire specimen was stored in 75% ethanol.
3.2 DiagnosisMacandrewia with contorted and undulating lamellae; desmas with short, rounded, blunt tubercles.
3.3 DescriptionSponge foliate with contorted and undulating lamellae (Fig. 2), approximately 7-cm high, 9-14-cm wide at the top, lamellae 4-mm thickness. Concave (upper) surface smooth and many small granular bumps is visible by naked eyes, and many microxeas protruded out on the surface shown in SEM (Fig. 3a), about 200 μm in diameter. Lower surface is smooth with several little openings invisible by naked eye, about 20-120 μm in diameter (Fig. 3b). Texture is stony. Color is brownish in alcohol.
Ectosomal skeleton comprises of a layer of overlapping, strongly incised and dentate phyllotriaenes, and abundant microxeas (Fig. 3a-b). Choanosome composed of an irregular reticulation of tuberculate and compact desmas accompanied by oxeas (Fig. 3c). Desmas (Fig. 3c-d) are strongly tuberculated tetraclones with four rays emanating from the center. Most of these desmas have triaenose crepis of which one of the four rays are longer than the rest.
Megascleres comprise of phyllotriaenes and desmas. Phyllotriaenes (Fig. 4b-c) have short conical rhabdome (77-94-117 μm in length and 17-22- 26 μm in width; presented as minimum-mean- maximum, the same afterward), broad dentate, and heavily incised cladome (186-237-313 μm in diameter) with strongly indented edges. Desmas have smooth clones and tubercles (Fig. 4c). Desmas overall sizes are 252-389-520 μm×32-42-61 μm in L×W manner, the same afterward. The desma clones, 78- 134-246 μm×21-27-32 μm in size, are branched at the end into many short, rounded, and blunt tubercles that articulate with each other into complex and tuberculated zygoses (Fig. 3e-f). Oxeas (Fig. 4a), slightly curved, are 256-382-476 μm×6-8-9 μm.
Microscleres comprise of only microxeas that are curved and their tips are blunt (Fig. 4e), 51-65- 91 μm×2-3-4 μm in size.
3.4 EtymologyNamed after the Yap Trench where the specimen was found in proximity.
3.5 Type localityA seamount near Yap Trench (Western Pacific), 255-311 m.
3.6 Substrate and ecologyThe sponge was attached to the stony oculinid coral, Cladopsammia sp., and flanked by a globular species of the sponge family Geodiidae Gray, 1867 both sitting on sandy substrate around 255-311 m (Fig. 2a).
3.7 RemarkMacandrewia yapensis sp. nov. possesses ectosomal phyllotriaenes and desmas with triaenose (rarely monaxial) crepis, smooth oxeas, and microxeas, which is key character of Macandrewiidae. Nine species of Macandrewia are known globally and only two previously known in the Pacific Ocean (Table 3). The new species clearly differs from the other two species by its foliate body shape with contorted lamellae, which differed from M. rigida from the Philippines that is a small and branching sponge not more than 45 mm in height and from M. spinifoliata from New Caledonia and New Zealand that is arbuscular branched or nubbin-shaped. In addition, the new species can be differentiated from M. rigida by having bigger phyllotriaenes and smaller oxeas, and can be distinguished from M. spinifoliata with its much larger desmas.
It seems that external morphology is the key distinguishing character in the genus Macandrewia: M. auris (earlobe-shaped), M. azorica (cyathiform to flabellate), M. clavatella (obconic), M. minima (globular), M. ramosa (encrusting), M. rigida (branching), M. robusta (foliate to globular), M. schusterae (foliate with contorted lamellas), and M. spinifoliata (branched). Only M. schusterae from the Atlantic Ocean share the same shape of foliate with thick and contorted lamellas with M. yapensis sp. nov. among the congeners. However, M. yapensis sp. nov. has tuberculate desma clones while M. schusterae has narrow and mostly pointed clones.
The tuberculiform clone tips of desmas may be an important distinguishable character of the new species as well. Macandrewia azorica, M. robusta, M. schusterae, and M. minima have short branching clones with blunt tips but the details of desmas of M. spinifoliata, M. clavatella, M. ramosa, M. rigida, and M. auris are unknown. The features at the terminal end of clones can be an important character among Macandrewia species.
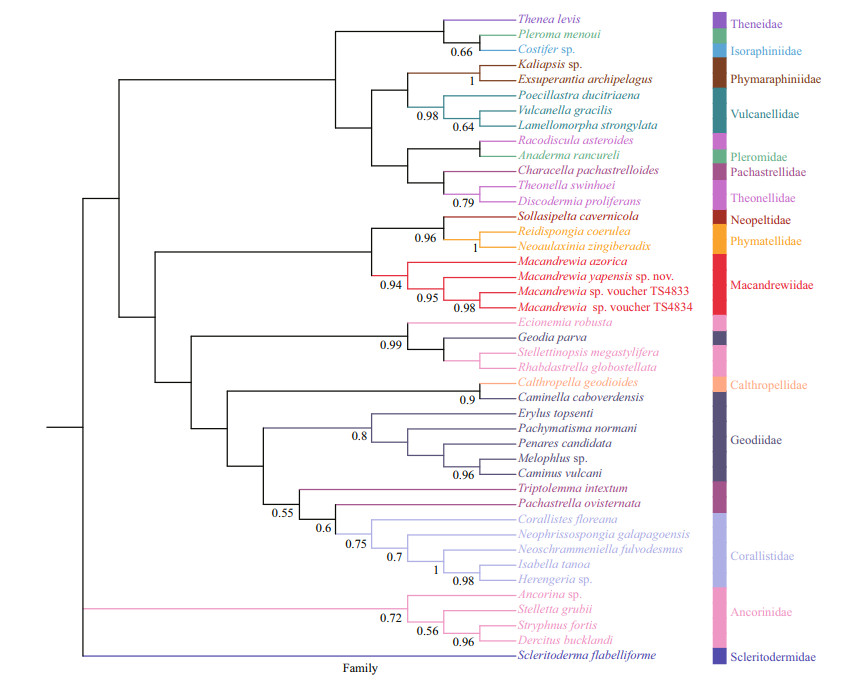
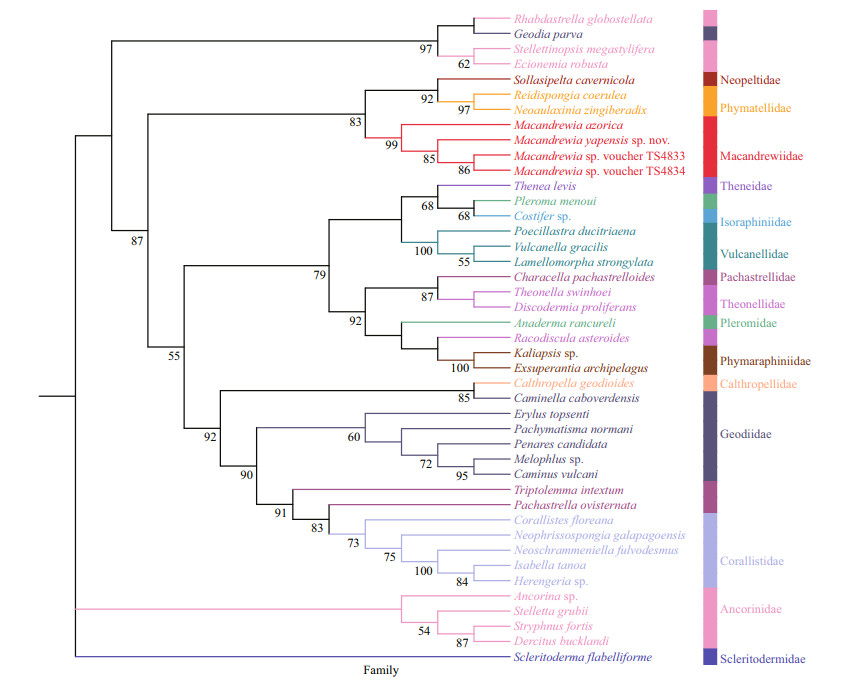
3.8 Molecular dataThe phylogenetic trees of BI (Fig. 5) and ML analyses (Fig. 6) are generally congruent. The new species and the three other Macandrewia species form a clade within the family Macandrewiidae, thus supporting the family assignment, with support values > 0.8 for both the ML and BI analyses.
|
| Fig.5 Phylogenetic tree obtained by the Bayesian inference analysis based on COI gene sequences Numbers at each node are Bayesian posterior probabilities (PP). Only values 0.5 are shown. |
|
| Fig.6 Phylogenetic tree obtained by ML analysis based on COI gene sequences Numbers at each node are maximum likelihood bootstrap values (BP). Only values 50% are shown. |
The phylogeny of Macandrewiidae based on morphology has been revised for many times (Kelly, 2000; Pisera and Lévi, 2002; Schuster et al., 2015). In addition, the phylogeny of the family has been explored through molecular approaches recently. Schuster et al. (2015) amplified the first 28S gene sequence of Macandrewia species (that is M. rigida), and it clustered with the genus of Calthropella Sollas, 1888 (family Calthropellidae Lendenfeld, 1907) and the subfamily Erylinae Sollas, 1888 with low support values (posterior probabilities (PP) < 0.75; bootstrap values (BP) < 60%). Schuster et al. (2021) amplified another 28S gene sequence of an undescribed Macandrewia sp., and it clustered with M. rigida initially. After that, the two macandrewiid species grouped with the genus of Calthropella and Caminella Lendenfeld, 1894 (subfamily Erylinae), both of them are non-desma bearing demosponges. The molecular approaches of Macandrewiidae based on 28S gene sequence are not consistent with the morphology approaches. In our phylogenetic trees based on COI sequences, Macandrewiidae is a sister with Neopeltidae and Phymatellidae clades that have desmas as well. It is consistent with the morphology approaches especially the results of Kelly (2000) who thought Macandrewia should belong to Neopeltidae. In our phylogenetic tree, Macandrewia species are not clustered with Neopeltidae species, which is consistent with the idea of Pisera and Lévi (2002) that Macandrewiabelongs to the family of Macandrewiidae but not Neopeltidae.
Comparing the phylogenetic tree constructed with 28S rDNA gene by Schuster et al. (2015) and Schuster et al. (2021), it seems that the COI gene tree is more congruent to the morphological data due probably to the low variation within the 28S rDNA gene (Schuster et al., 2015). The results of our analysis did not contain all sequences of astrophorins, and more molecular markers and sequences are required in the future to build a more comprehensive tree by which the systematic of astrophorins will be elucidated to a detail.
5 DATA AVAILABILITY STATEMENTThe authors declare that the data supporting the findings of this study are available within the article. The original data of spicule measurements will be available on request from the first author.
6 ACKNOWLEDGMENTWe are grateful to Dr. Kuidong XU (Institute of Oceanology, Chinese Academy of Sciences, Qingdao) for providing the deep-sea sponge specimens. Special thanks to the reviewers for helping us improve the manuscript.
Bergquist P R, Hogg J J. 1969. Free amino acid patterns in Demospongiae: a biochemical approach to sponge classification. Cahiers de Biologie Marine, 10(2): 105-220.
|
Carvalho F C, Cárdenas P, Ríos P, Cristobo J, Rapp H T, Xavier J R. 2020. Rock sponges (lithistid Demospongiae) of the Northeast Atlantic seamounts, with description of ten new species. PeerJ, 8: e8703.
DOI:10.7717/peerj.8703 |
Edgar R C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32(5): 1 792-1 797.
DOI:10.1093/nar/gkh340 |
Gray J E. 1859. Description of MacAndrewia and Myliusia, two new forms of Sponges. Proceedings of the Zoological Society of London, 27: 437-440.
DOI:10.1111/j.1469-7998.1859.tb00355.x |
Huelsenbeck J P, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics, 17(8): 754-755.
DOI:10.1093/bioinformatics/17.8.754 |
Kelly M. 2000. Description of a new lithistid sponge from northeastern New Zealand, and consideration of the phylogenetic affinities of families Corallistidae and Neopeltidae. Zoosystema, 22(2): 265-283.
|
Kelly M. 2007. The marine fauna of New Zealand: Porifera: lithistid Demospongiae (rock sponges). NIWA Biodiversity Memoirs, 121: 1-100.
|
von Lendenfeld R.. 1907. Die Tetraxonia. Wissenschaftliche Ergebnisse der Deutschen Tiefsee—Expedition auf der Dampfer Valdivia 1898-1899. Jena, 11(1-2): 59-374.
|
Letunic I, Bork P. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Research, 47(W1): W256-W259.
DOI:10.1093/nar/gkz239 |
Lévi C, Lévi P. 1983. Eponges Tétractinellides et Lithistides bathyales de Nouvelle-Calédonie. Bulletin du Muséum national d'Histoire naturelle, 5(1): 101-168.
|
Lévi C, Lévi P. 1989. Spongiaires (MUSORSTOM 1 & 2). In: Forest J ed. Résultats des Campagnes MUSORSTOM, volum 4. Mémoires du Muséum national d'Histoire naturelle, Paris, Fanch. p. 48-49.
|
Marshall W. 1876. Ideen über die Verwandtschaftsverhältnisse der Hexactinelliden. Zeitschrift für wissenschaftliche Zoologie, 27(1): 113-136.
|
Meyer C P, Geller J B, Paulay G. 2005. Fine scale endemism on coral reefs: archipelagic differentiation in turbinid gastropods. Evolution, 59(1): 113-125.
DOI:10.1111/j.0014-3820.2005.tb00899.x |
Morrow C, Cárdenas P. 2015. Proposal for a revised classification of the Demospongiae (Porifera). Frontiers in Zoology, 12: 7.
DOI:10.1186/s12983-015-0099-8 |
Pisera A, Lévi C. 2002. Family Macandrewiidae Schrammen, 1924. In: Hooper J N A, van Soest R W M, Willenz P eds. Systema Porifera: A Guide to the Classification of Sponges. Springer, Boston, MA. p. 377-379, https://doi.org/10.1007/978-1-4615-0747-5_45.
|
Posada D, Crandall K A. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics, 14(9): 817-818.
DOI:10.1093/bioinformatics/14.9.817 |
Rambaut A, Drummond A J, Xie D, Baele G, Suchard M A. 2018. Posterior summarization in Bayesian phylogenetics using tracer 1.7. Systematic Biology, 67(5): 901-904.
DOI:10.1093/sysbio/syy032 |
Schlitzer R. 2020. Ocean Data View, https://odv.awi.de.
|
Schmidt O. 1870. Grundzüge einer Spongien-Fauna des atlantischen Gebietes. Verlag von Wilhelm Engelmann, Leipzig, Germany. p. 23-24.
|
Schrammen A. 1924. Die Kieselspongien der oberen Kreide von Nordwestdeutschland. III. und letzter Teil. Monographien zur Geologie und Paläontologie, 1: 1-159.
|
Schuster A, Erpenbeck D, Pisera A, Hooper J, Bryce M, Fromont J, Wörheide G. 2015. Deceptive desmas: molecular phylogenetics suggests a new classification and uncovers convergent evolution of lithistid demosponges. PLoS One, 10(1): e116038.
DOI:10.1371/journal.pone.0116038 |
Schuster A, Pomponi S A, Pisera A, Cárdenas P, Kelly M, Wörheide G, Erpenbeck D. 2021. Systematics of 'lithistid' tetractinellid demosponges from the Tropical Western Atlantic—implications for phylodiversity and bathymetric distribution. PeerJ, 9: e10775.
DOI:10.7717/peerj.10775 |
Sollas W J. 1885. A classification of the sponges. Annals and Magazine of Natural History, 16(95): 395.
|
Sollas W J. 1887. Sponges. In: Black A C ed. Encyclopaedia Britannica, 9th edition. Volume 22. Encyclopaedia Britannica, Edinburgh, Scotland. p. 412-429.
|
Sollas W J. 1888. Report on the Tetractinellida collected by H.M.S. Challenger, during the years 1873-1876. Report on the Scientific Results of the Voyage of H.M.S. Challenger during the years 1873-76. Zoology, 25: 1-458.
|
Topsent é. 1904. Spongiaires des Açores. Résultats des campagnes scientifiques accomplies par le Prince Albert I. Monaco, 25: 1-280.
|
Trifinopoulos J, Nguyen L T, von Haeseler A, Minh B Q. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Research, 44(W1): W232-W235.
DOI:10.1093/nar/gkw256 |
Van Soest R W M, Boury-Esnault N, Hooper J N A, Rützler K, de Voogd N J, Alvarez B, Hajdu E, Pisera A B, Manconi R, Schönberg C, Klautau M, Kelly M, Vacelet J, Dohrmann M, Díaz M C, Cárdenas P, Carballo J L, Ríos P, Downey R, Morrow C C. 2021. World Porifera Database. Macandrewiidae Schrammen, 1924. http://www.marinespecies.org/porifera/porifera.php?p=taxdetails& id=171122. Accessed on 2021-01-18.
|
Zhang D, Gao F L, Jakovlić I, Zou H, Zhang J, Li W X, Wang G T. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Molecular Ecology Resources, 20(1): 348-355.
DOI:10.1111/1755-0998.13096 |
 2021, Vol. 39
2021, Vol. 39