Institute of Oceanology, Chinese Academy of Sciences
Article Information
- HAN Lingshu, QUAN Zijiao, HAN Bing, DING Beichen, HUANG Xiaofang, WANG Heng, CHANG Yaqing, DING Jun
- Molecular characterization and expression of the SiUCP2 gene in sea urchin Strongylocentrotus intermedius
- Journal of Oceanology and Limnology, 39(4): 1523-1537
- http://dx.doi.org/10.1007/s00343-020-0181-8
Article History
- Received May. 8, 2020
- accepted in principle Jul. 3, 2020
- accepted for publication Sep. 1, 2020
Adenosine triphosphate (ATP) represents one of the final compounds produced by animal organisms to store chemical energy converted from food. The generation of ATP is related to the energy requirements of the organism and is closely related to the metabolic rate. The metabolic rate quantifies the energy requirements of the organism, which is related to the size of the animal; therefore, the larger the body size is, the higher the metabolic rate is (McCGraham, 1967). The metabolic rate is always in a state of fluctuation, which varies with the amount of energy used by the animal. When the animal is in a stage of reproduction or stress, the metabolic rate would change accordingly. Most organisms maintain a basal metabolic rate (BMR) before they consume additional energy for life activities. Therefore, organisms have evolved a feedback system called "energy balance" that responds to energy requirements, so that the metabolic rate can change with energy requirements (Campbell and Reece, 2005).
When starvation occurs, there lacks sufficient energy substances to meet the needs of the juvenile perch (Perca fluviatilis), thus the organism first uses glycogen stored in the body, and then uses fat as energy substance, which is oxidized and decomposed to maintain energy metabolism, and finally the organism uses protein (Mehner and Wieser, 1994). However, some aquatic animals use energy substances in a different order during starvation. Fish Sciaenops ocellatus (Jiang et al., 2002) and juvenile Eriocheir sinensis (Wen et al., 2002) prioritize the use of fats and carbohydrates, while a few species of fish, such as Pagrus major (Zhang et al., 2000) mainly use protein as an energy source during starvation. Fatty acids (FAs) are an important source of energy for cells and are the primary raw material for triglyceride synthesis. Polyunsaturated fatty acids (PUFAs) play important functional roles in a variety of tissues, such as promoting nervous system development, mediating immunity, and promoting coagulation processes. Fatty acid derivatives and metabolites are involved in mediating multiple gene expression and enzymatic reactions, ion channel and membrane receptor activities, etc. FA metabolism in animals is a complex process that is regulated by several factors such as genetics, hormones, nutrition, and immunity. Factors such as inheritance influences fat deposition (Geri et al., 1990). Fat deposition depends on the effects of several enzymes during fat synthesis and catabolism, as well as the regulation of hormones involved in fat metabolism (Malavazos et al., 2007; Jiang et al., 2008; Kang et al., 2017). Researchers have undertaken several studies on fat deposition and its regulatory mechanism in livestock for many years, and found that candidate genes such as UCP1, UCP2, UCP3, LPL, PPARs, Leptin, and SREBP were closely associated with fat metabolism and meat quality (Douaire et al., 1992; Taouis et al., 1998; Huang and Xie, 2004; Li et al., 2010).
Uncoupling proteins 2 (UCP2) belongs to UCPs family which is a class of proton transporters located in the inner mitochondrial membrane and are members of the mitochondrial anion carrier superfamily (MACF) (Fleury and Sanchis, 1999; LuévanoMartínez, 2012). Studies have shown that UCP2 is involved in multiple metabolic processes such as food intake (Coppola et al., 2007; Andrews et al., 2008), FA metabolism (Klingenberg and Echtay, 2001; Brookes et al., 2008), insulin secretion (Wang et al., 2010), and the immune response (Tagen et al., 2009; Emre and Nübel, 2010). Currently, the UCP2 gene has been cloned from rat (Matsuda et al., 1997), pig (Werner et al., 1999), carp and zebrafish (Stuart et al., 1999), and silver carp (Liao et al., 2006), but the cloning of UCP2 from Echinoderms has not yet been reported.
Strongylocentrotus intermedius was introduced to China from Japan by Dalian Ocean University in 1989 and is currently distributed in Liaoning and Shandong Peninsula. It is one of the main species of sea urchins cultured in China (Chang et al., 2004). The gonads of sea urchins are the only edible part of the organism. These organs contain essential nutrients such as lipids and PUFAs, which not only determine the nutritional value of sea urchins, but also ensure the normal growth and reproduction of the organisms during cultivation processes. Uneven feeding or excessive feeding often happens in the aquaculture of sea urchins. Moderate deposits of fat help the sea urchins maintain normal metabolic activities, but excessive deposits will not only affect the health of the sea urchins but also reduce the quality of the gonads (Zhou et al., 2008). Artificially cultured sea urchins are often exposed to starvation stress due to environmental and seasonal changes. During starvation, sea urchins will consume stored glycogen and fat to maintain their basal metabolism (Lares and Pomory, 1998). UCP2 regulates lipid metabolism directly or indirectly, but the structure and expression of UCP2 and its role in lipid metabolism in S. intermedius have not been reported.
In this study, the SiUCP2 gene was cloned from S. intermedius using the rapid-amplification of cDNA ends (RACE) technique, and the expressions of this gene in different tissues and at different development stages and in different starvation periods of S. intermedius were studied using quantitative realtime PCR (qRT-PCR). Gene expression in intestinal and gonadal tissues was also examined by in-situ hybridization. Finally, the expression of SiUCP2 recombinant protein in the intestine and gonads were examined using western blotting. This study aimed to investigate the structure and function of the SiUCP2 gene from S. intermedius and its expression at the mRNA and protein level to provide a theoretical basis for subsequent studies on the role of SiUCP2 and its regulatory mechanism the fatty acids synthesis of S. intermedius. In the future, we will continue to pay attention to SiUCP2, study which fatty acids are produced by SiUCP2, clarify the mechanism of SiUCP2 in the synthesis of fatty acids, improve the content of fatty acids in sea urchins, and further improve the gonadal quality of sea urchins to obtain sea urchins with a higher economic value.
2 MATERIAL AND METHODSea urchins and rabbits used in this study were farmed, and all experiments were conducted in accordance with the ethics committee of Dalian Ocean University and national guidelines. No endangered or protected species were involved in this test. The location of the training experiment does not require special permission.
2.1 Animals, rearing conditions, and sample collectionOne-year-old S. intermedius adults used in this study were bred at Dalian Ocean University. Healthy sea urchins were homogeneous in size with test heights of 21.80±1.20 mm, shell lengths of 37.62±1.53 mm, and weights of 24.27±1.46 g.
The tube feet, coelomocytes, Aristotle's lantern, stomachs, and gonads, intestines, were dissected from three sea urchins. To collect samples from different developmental stages, we selected three females and three males and injected with 1-mL KCL (40 μL/g of body mass) to induce spawning. Unfertilized eggs were transferred to a beaker containing 1.5-L fresh sea water (salinity: 31) in density of 20–30 ind./mL in temperature of 19±0.5 ℃. They were then dripped into semen (sperm-egg ratio was 1 000∶1). Water was stirred to ensure sperm and eggs in good contact, and stayed for 15 min. After artificial fertilization, eggs were washed three times every 30 min to remove extra sperm and impurities. Morphological changes in larvae were observed under a microscope (Kelly et al., 2000). The fertilized eggs develop in three stages, early embryonic development, planktonic larva, and competent larvae, and four major developmental stages, the blastula, prismatic larval, long-arm larval, and juvenile sea urchin. All samples were frozen in liquid nitrogen immediately and stored at -80 ℃.
A starvation experiment commenced in May 2019, in which 25 sea urchins were cultured in a 500-L tank in water temperature of 19 ℃. Feces were observed every day since feeding was stopped. Once fecal excretion was significantly reduced (Li et al., 2004), five samples were taken as the control group, and the hunger experiment officially commenced. The intestines and gonads of five sea urchins were taken on Days 0, 7, 14, and 21 of starvation and rapidly frozen in liquid nitrogen, and stored at -80 ℃.
2.2 Total RNA extraction and reverse transcriptionTotal RNA was extracted from different tissues, at different development stages and different starvation periods of S. intermedius, using an RNAprep Pure Kit for tissue (Tiangen Biotech, Beijing, China) according to the manufacturer's protocol. Standard RNAs were used for cDNA preparation. The cDNA for qRT-PCR was prepared using a Prime ScriptTM RT Reagent Kit (TaKaRa, Japan). The reaction was performed in a total volume of 10 μL containing 2-μL 5×Primer script Buffer, 0.5-μL Primer script Enzyme MixⅠ, 0.5- μL Oligo dT Primer, 0.5-μL Random Primer, 500-ng Total RNA and ddH2O. The PCR reaction conditions were 37 ℃ for 15 min and 85 ℃ for 5 s. All cDNA samples were diluted five times and stored at -20 ℃ before subsequent experiments.
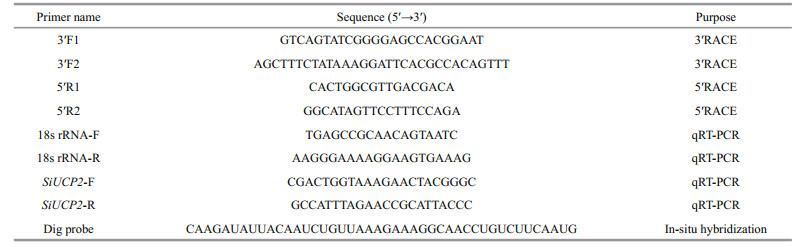
2.3 Cloning the full-length cDNA of SiUCP2 using rapid amplification of cDNA ends (RACE) PCRThe partial cDNA sequence of SiUCP2 was obtained from our transcriptome assembly data (http://gsa.big.ac.cn/, accession number: PRJCA000276), and the gene-specific primers were designed by Primer 5.0 and obtained from Sangon Biotech, Shanghai as shown in Table 1. The cDNA for RACE was prepared using the M-MuLV First Strand cDNA Synthesis Kit (Sangon, Shanghai, China). The PCR reactions for 5′-RACE and 3′-RACE were conducted following the manufacturer's instructions. Products from RACE were extracted and purified using a Quick Gel Extraction kit (Transgen Biotech, China). For specific experimental content, refer to kits instructions and Han et al. (2019). After the fulllength cDNA of SiUCP2 was amplified, it was ligated into the PEASY-1 vector and sequenced.
The open reading frame (ORF) of SiUCP2 was analyzed using ORF Finder (https://www.ncbi.nlm. nih.gov/orffinder/) and the deduced amino acid (AA) sequence was determined using the Expert Protein Analysis System (http://www.expasy.org/). The physical and chemical parameters of the deduced protein were computed by the ProtParam tool (http://web.expasy.org/protparam/). The functional domains of the SiUCP2 protein were predicted by InterPro (http://www.ebi.ac.uk/interpro/). Multiple sequence alignment of the deduced SiUCP2 AA sequence was performed using DNAMAN 6.0. The phylogenetic tree was constructed using the neighbor-joining method in MEGA 7.0.
2.5 Real-time PCR analyses of SiUCP2 expressionThe relative expression levels of SiUCP2 mRNA transcripts in different tissues (coelomocytes, tube feet, Aristotle's lantern, gonads, intestines and stomachs), were assessed at developmental stages (eggs (1–2 min after fertilization), two cells (40 min after fertilization), eight cells (2 h after fertilization), blastula (5–6 h after fertilization), gastrula (17–20 h after fertilization), prism larvae (24 h after fertilization), two-arm larvae (30–40 h after fertilization), eight-arm larvae (7–13 d after fertilization), juvenile sea urchins (30 d after fertilization)) and intestines and gonads of sea urchins that were fasted for 0, 7, 14, and 21 d were analyzed with quantitative real-time PCR (qRT-PCR) using the Applied Biosystems 7500 Real-time System (Applied Biosystems, Foster City, CA, USA) and SYBR Premix Ex Taq (SYBR PrimeScriptTM RT-PCR kit II, TaKaRa, Japan) following the manufacturer's instructions. The 18s rRNA gene was used as an internal reference gene. The amplification was performed in a total volume of 20 μL containing 2 μL of 1∶5 diluted original cDNA, 10 μL of 2×SYBR Green Master mix (TaKaRa, Japan), 0.4 μL of ROX Reference Dye, 6 μL of PCR grade water and 0.8 μL (10 mmol/L) of each primer. The reaction conditions were followed by 40 cycles of 94 ℃ for 5 min, 94 ℃ for 30 s, 60 ℃ for 30 s, and 72 ℃ for 30 s. The final extension step was at 72 ℃ for 5 min. Three independent biological replicates and three technical repetitions of each group were carried out. The relative expression levels of the target gene were calculated by the 2-ΔΔCt method (Livak and Schmittgen, 2001).
2.6 Statistical analysisAll data were expressed as the mean±SD (n=5). P value was adjusted for multiple tests using a false discovery rate (Benjamini-Hochberg). Significant differences (P < 0.05) for each variable were firstly detected using the one-way ANOVA test, followed by Tukey's test and Tamhane's T2 method. Statistical analysis was performed using SPSS software version 19.0 (IBM, Armonk, NY, USA).
2.7 In-situ hybridizationIn-situ hybridization was performed using a DIG High Prime DNA Labeling and Detection Starter Kit I (Sangon). Probe primers were based on the full-length cDNA sequence of SiUCP2. The intestines and gonads of S. intermedius were collected at the three stage of reproductive cycle the gonads and soaked in a 4% paraformaldehyde solution overnight. After the tissues were fixed, dehydrated with gradient alcohol, waxed, and embedded, they were cut into slices and baked in oven at 62 ℃ for 2 h. Paraffin sections were dewaxed by placing in water and sequentially in xylene I for 15 min, xylene II for 15 min, anhydrous ethanol I for 5 min, anhydrous ethanol II for 5 min, 85% alcohol for 5 min, 75% alcohol for 5 min, and washed in diethylpyrocarbonate (DEPC). Slices were placed in the repair solution, boiled for 10–15 min, and cooled naturally. Proteinase K (20 μg/mL) was added over 30 min at 37 ℃ and washed with pure water, the prehybrid solution was added and incubated at 37 ℃ for 1 h and then decanted. The hybridization solution containing the probe was added at a concentration of 5 ng/μL, and the mixture was mixed overnight at 37 ℃ in an incubator. To wash away the hybridization solution, the sections were washed by 2×saline sodium citrate (SSC) at 37 ℃ for 10 min, washed by 1×SSC at 37 ℃ for 5 min, and finally washed by 0.5×SSC at room temperature for 10 min. Serum bovine serum albumin (BSA) was added and incubated at room temperature for 30 min. After pouring off the sealing solution, anti-digoxin-labeled alkaline phosphatase was added and incubated at 37 ℃ for 40 min. Then washed for 5 min with phosphate buffer saline (PBS) for four times. After the sections were slightly dried, the freshly prepared nitro-blue tetrazolium (NBT) color rendering solution was added, and the color rendering time was controlled under the microscope. The sections were washed with pure water to terminate the color rendering, and then nuclear solid red dye was added to stain the nucleus, which was washed with pure water after an appropriate degree of staining. Finally, neutral gum was used to seal the film and an optical microscope was used to observe the results.
2.8 Construction of prokaryotic expression plasmid and preparation of antibody 2.8.1 Construction of prokaryotic expression plasmidThe encoding region of SiUCP2 was amplified by a gene-specific primer with the enzyme cleavage site Nde I/Xho I. After sequencing, it was inserted into the expression vector pET22b (Sangon) to construct the recombinant plasmid pET22b-SiUCP2, which was expressed in Escherichia coli.
2.8.2 Animal immunityFour-month-old healthy female New Zealand white rabbits weighing 2.1 kg were used. For the first immunization, the protein antigen was emulsified with an equal volume of Freund's complete adjuvant and injected (100 μL) into the lymph nodes of the hind legs. A second immunization was performed after 21 d. The protein antigen was emulsified with an equal volume of Freund's incomplete adjuvant and injected (100 μL) into the lymph nodes of the hind legs. The third immunization was performed after 35 d. After 42 d, 1 mL of blood was collected from the ear vein and the antiserum titer was detected by enzyme-linked immunosorbent assay (ELISA).
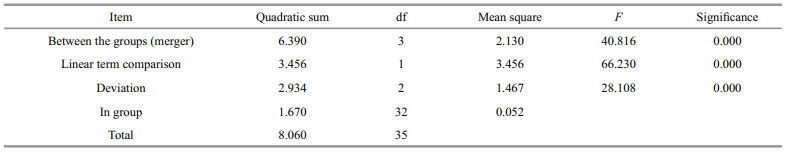
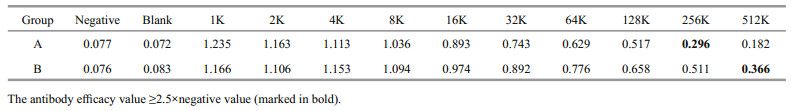
2.8.3 Indirect ELISA testThe antigen was coated with 0.05-mol/L carbonate (pH=9.6) at 100 μL/well, incubated at 4 ℃ overnight, removed and washed for 3 min with 0.05% Tween-20 (PBST) for three times. Blocking solution (150 μL of 5% skimmed milk) was added to each well and blocked at 37 ℃ for 60 min, and then washed for 3 min three times with 0.05% Tween-20 (PBST). The rabbit serum was diluted 1∶1 000 and then incubated at 37 ℃ for 1 h, and washed three times with 0.05% Tween-20 (PBST) for 3 min per time. Horseradishlabeled enzyme goat anti-rabbit IgG (H+L) (Sangon) was diluted 1∶8 000 and incubated at 37 ℃ for 45 min. The plate was washed five times with 0.05% Tween-20 (PBST) and 100 μL/well of substrate solution (TMB) was added, the reaction was carried out for 15 min. Finally, 100 μL of 2-mol/L sulfuric acid was added to terminate the reaction. Optical density (OD) value was measured at a wavelength of 450 nm using a microplate reader (KeHua ST-360).
2.9 Extraction and expression of the SiUCP2 proteinAbout 20-mg gonads were taken and 200-L RIPA lysate was added (with 1-mmol/L PMSF, broadspectrum protease inhibitor, and phosphatase inhibitor). Low temperature and high speed homogenate for 3 times, 10 s each time, 30 s pause, then ice lysis for 2 h, and take out ice bath ultrasound once in the middle. At 4 ℃, 12 000×g high speed centrifugation was performed, the supernatant was absorbed and centrifuged again, and the intermediate clarified sample was taken again until the obtained protein solution was clarified and transparent, which was temporarily stored at 4 ℃. The protein concentration was determined. In this study, western blotting was used to detect SiUCP2 in the gonads of S. intermedius, with an antibody that was diluted 1∶2 000 and β-actin acted as a house-keeping gene. The resultant protein was isolated on a reduced sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE, 10% separating gel and 5% stacking gel, 40-μg protein extract was loaded into lanes for electrophoresis) and visualized after staining with Coomassie Brilliant Blue R-250.
3 RESULT 3.1 Analysis of SiUCP2 sequenceThe SiUCP2 gene was obtained from S. intermedius by RACE technology and the sequence was deposited in the GenBank database, under the accession number MN065154. SiUCP2 was 2 341 bp in length and contained a 969-bp ORF encoding 323 AAs, a 232-bp 5′UTR, and a 1 140-bp 3′UTR containing a polyadenylation signal (AATAAAA). The molecular weight of SiUCP2 was 36.11 kDa, and the theoretical pI was 9.68. Analysis of the AA sequence predicted that SiUCP2 had no signal peptide and three characteristic features of carrier proteins in the inner mitochondrial membrane. Analysis of the AA hydrophilicity/hudrophobicity indicated that SiUCP2 had an aliphatic index of 87.24 and a grand average of hydropathicity (GRAVY) of 0.014, thus SiUCP2 was a hydrophobic protein (Fig. 1).

|
| Fig.1 Nucleic acid sequence of SiUCP2 in S. intermedius and its deduced amino acid sequence The starting codon ATG is indicated in bold, the termination codon TAA is indicated by bold italics and the box contains the characteristic domain of the carrier protein in the mitochondrial inner membrane. The shaded region represents the characteristic sequence of uncoupled proteins, and the underlined region represents the polyA tail signal (AATAAAA). |
The AA sequence of SiUCP2 was aligned with UCP2 AA sequences from other species using DNAMAN 6.0 software. The results showed that SiUCP2 shared a high homology with all UCP2 from vertebrates and invertebrates as shown in Table 2.
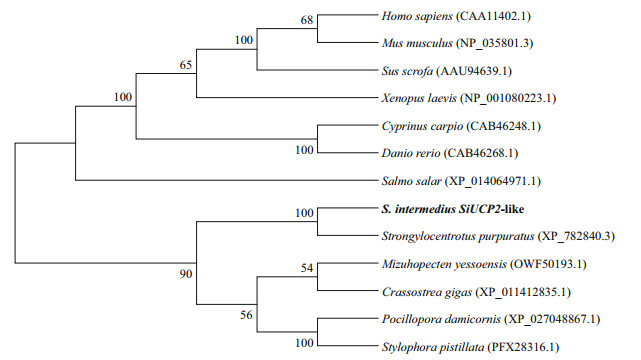
To reveal the phylogeny of the SiUCP2 cloned in this study, a phylogenetic tree was constructed using the neighbor-joining method. The result in Fig. 2 shows that SiUCP2 from S. intermedius and UCP2 from S. purpuratus were clustered into one branch, indicating that these two species were closely related.
|
| Fig.2 Consensus neighbor-joining tree based on the amino acid sequences of SiUCP2 genes from other species The evolutionary history was inferred using the neighbor-joining method. The percentage of replicate trees where the associated taxa cluster together in the bootstrap test (1 000 replicates) are shown next to the branches. Evolutionary analyses were conducted in MEGA 7.0. |
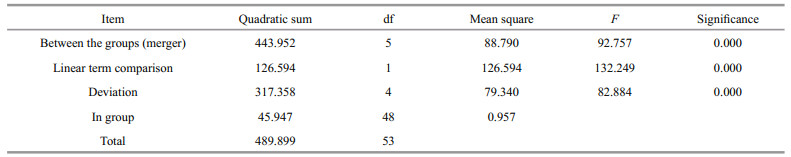
The tissue distribution of SiUCP2 mRNA in S. intermedius was investigated by qRT-PCR. The results in Fig. 3 show that the SiUCP2 gene was expressed in all tissues examined. The data analysis results are shown in Table 3. Descriptive statistical analysis is shown in Supplementary Table S1. The relative expression level was the highest in the tube feet, followed by the gonads, and the relative expression level was the lowest in the coelomocytes.

|
| Fig.3 Relative expression levels of SiUCP2 in different tissues Each vertical bar represents the mean±SD (n=5), 18s rRNA was used as a reference gene. Letters above the bars indicate significant differences at P < 0.05, in different tissues. |
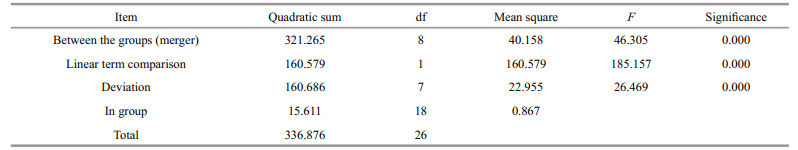
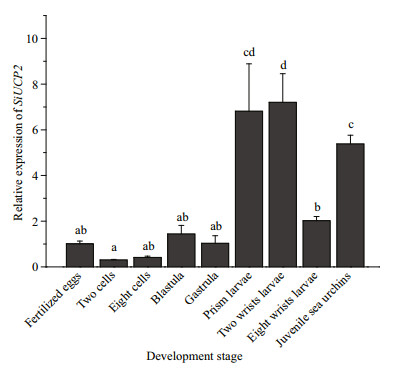
The expression of SiUCP2 in S. intermedius at different development stages, including fertilized eggs, two cells, eight cells, blastulae, gastrula, prism larvae, two-arm larvae, eight-arm larvae, and juvenile sea urchins, was determined to understand the role of the SiUCP2 gene in different development stages. As shown in Fig. 4, The data analysis results are shown in Table 4. Descriptive statistical analysis is shown in Supplementary Table S2. SiUCP2 was expressed at all nine development stages and the relative expression at the prism larval stage was the highest, while the expression at the 2-cell stage was significantly lower than at other development stages. During the whole development process, the overall expression of SiUCP2 showed a pattern of first increasing and then decreasing.
|
| Fig.4 Relative expressions of SiUCP2 during the development of S. intermedius 18s rRNA was used as a reference gene. Each vertical bar represents the mean±SD (n=3). Letters above the bars indicate significant differences at P < 0.05, in different development stages. |
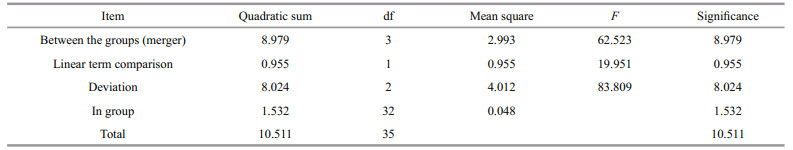
The relative expression of SiUCP2 in the intestines and gonads of S. intermedius at different starvation periods was determined using qRT-PCR to understand the expression and role of SiUCP2 during starvation. The data analysis results are shown in Tables 5–6. Descriptive statistical analysis is shown in Supplementary Tables S3–S4. As shown in Fig. 5, the trends of different periods of starvation on SiUCP2 expression in the intestines and gonads of S. intermedius were similar. The relative expression levels of SiUCP2 in the intestines and gonads during starvation showed a pattern of first decreasing, then increasing and then decreasing again.
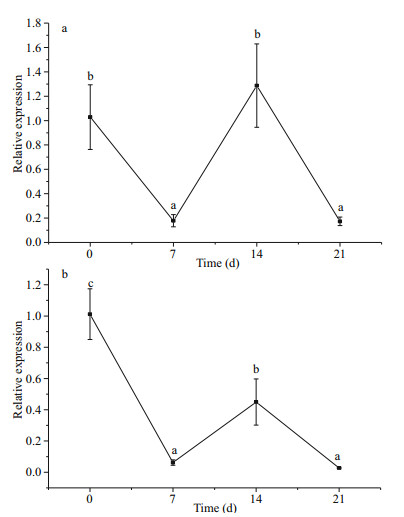
|
| Fig.5 Effects of starvation on the relative expression of SiUCP2 gene in gonads (a) and intestines (b) 18s rRNA was used as a reference gene. Each vertical bar represents the mean±SD (n=5). Different letters above the bars indicate significant differences at P < 0.05, at different times. |
In-situ hybridization of SiUCP2 in the intestines and gonads of S. intermedius was carried out. The results are shown in Fig. 6. Scattered blue-violet positive signals in the gonads were observed under a microscope. The signals were present in nutritive phagocytes, while no positive signals were observed in the intestines, indicating that SiUCP2 was expressed in small amounts in the gonads and intestines.
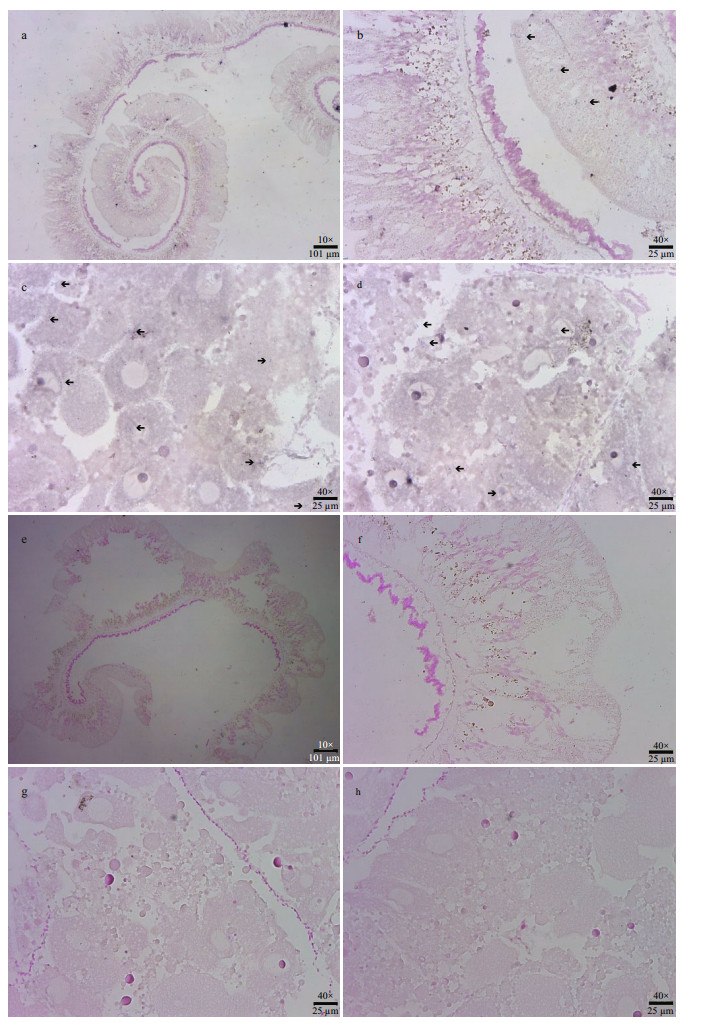
|
| Fig.6 In-situ hybridization of the SiUCP2 gene in intestines and gonads a & b. hybridization of the digoxigenin-labeled RNA probe with the sea urchin intestines tissue; c & d. the digoxigenin-labeled RNA probe hybridizes with the sea urchin gonad; e & f. hybridization of the sense probe with the sea urchin intestines; g & h. hybridization of the sense probe with the sea urchin gonads. The black arrow indicates a positive signal. Scales are shown in the figure. |
The prepared antibody was subjected to indirect ELISA. The OD values were measured using a microplate reader at 450-nm wavelength. The results are presented in Table 7.
To investigate the function of the SiUCP2 protein, total protein was extracted from the gonads of three sea urchins. The SiUCP2 protein was detected by western blotting using β-actin as an internal reference. Through the pre-experiment, we determined the destination band in the gonads, as shown in Fig. 7. The results of SDS-PAGE in Fig. 8 show that a clear band of about 36 kDa appeared in the gonads.
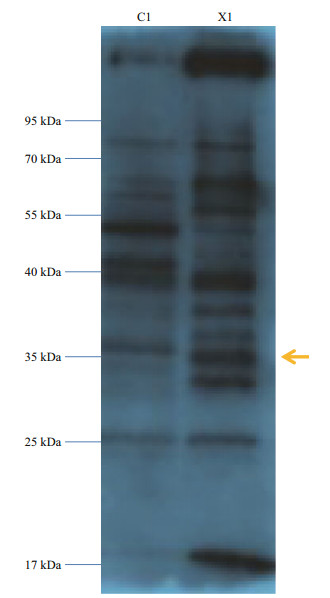
|
| Fig.7 Pre-experiment of expression of the SiUCP2 protein in the intestine and gonad of S. intermedius C1: the intestine; X1: the gonads; arrow: the destination band. |
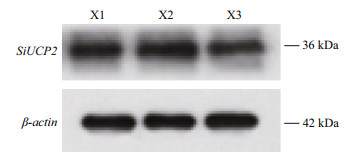
|
| Fig.8 Expression of the SiUCP2 protein in gonads of S. intermedius (n =3) X1, X2, and X3 represent the gonads of the three sea urchins. |
Sea urchins are considered delicacies in many countries. The gonads of sea urchins contain essential nutrients such as lipids and PUFAs, which not only determine the nutritional value of the sea urchins but also ensure the proton transporters, are located in the inner mitochondrial membrane. Free FAs provide necessary free carboxyl groups for UCPs, making proton transport possible or facilitating proton transport. Currently, the mechanisms of proton leaks caused by UCPs are still controversial (Garlid et al., 2000). UCPs that are present in the inner mitochondrial membrane are capable of disrupting the coupling between oxidation in the electron transport chain and adenosine triphosphate (ATP) synthesis, thus dissipating energy in the form of heat and possibly affecting metabolic efficiency (Thompson and Kim, 2004). The full-length cDNA of a gene is the basis for studying gene structure, function, and protein expression. The SiUCP2 gene of S. intermedius was characterized in this study. DNAMAN sequence alignment showed that SiUCP2 shared high homology with UCP2 from other species and had similar domains; therefore SiUCP2 may also have similar biological functions to other homologs from different species. Phylogenetic analysis showed that SiUCP2 from S. intermedius and UCP2 from S. purpuratus were clustered into one branch, indicating that these two species were closely related and distantly related to other vertebrates. Sequence similarity, conserved domains, and phylogenetic analysis provided evidence that the cloned gene was SiUCP2 cDNA.
UCP2 was highly expressed in the liver of Pagrus major, but almost undetectable in visceral mesenteric adipose tissue (Liang et al., 2003). The relative expression levels of UCP2 were high in the tissues of liver, stomach, and eye of Siniperca chuatsi, but low in the spleen, intestine, and brain (Wen et al., 2002). The qRT-PCR analysis showed that the SiUCP2 gene was expressed in all examined tissues of S. intermedius, which was consistent with the study results in other species (Wen et al., 2002; Coulibaly et al., 2006; Liao et al., 2006). Sea urchin tube feet, which facilitate movement and adhesion, contain a large amount of inorganic residues (45.5%), protein (6.4%), lipid (2.5%), and neutral sugar (1.2%) (Santos et al., 2009). The highest relative expression level of SiUCP2 in tube feet could be related to the energy consumption during movement and adhesion, which will be investigated in a follow-up study.
The high relative expression level of SiUCP2 in gonads may be due to fat metabolism in the gonads. The gonads of sea urchins store fat and are the main sites for fat metabolism. The decomposition of triglycerides produces a large amount of free FAs, resulting in a high expression level of SiUCP2 in the gonads, which was consistent with previous studies (Medvedev et al., 2002; Li et al., 2010). Sea urchin gonads contain nutritive phagocytes (NPs), which are versatile somatic cells that provide structural and nutritional microenvironments for germinal cells throughout sea urchin gametogenesis. In addition to mobilizing stored nutrients, NPs also continue to accumulate additional nutrients. NPs may also devour remaining eggs or sperm, to simply recover nutrients (Walker et al., 2013). SiUCP2 may be expressed in NPs and participate in nutrient metabolism. Sea urchins feed mainly on large algae such as kelps, Wakame, and sea lettuces. Some algae contain toxins, and reactive oxygen species (ROS) are inevitably produced during the oxidation of toxins in sea urchins. Echtay et al. (2002) found that peroxides promoted UCP2 gene expression, while a high expression of UCP2 effectively inhibited overproduction. The pharynx of the sea urchin is contained in Aristotles' lantern. The pharynx, stomach, and intestine constitute the digestive tract of the sea urchin. It is an important place for digestion and absorption in the body. The digestive tract has a certain fat content and immune functions, which prevent pathogenic and poison invasion. Algae food with certain toxins enter the gastrointestinal tract through the mouth to be digested, and the coelomic fluid of sea urchins is not only involved in transport, secretion, and buffering but, more importantly, is involved in immune defense. Some studies have confirmed that amebocyte in sea urchin coelomic fluid play a role in phagocytosis to kill non-self-substances and mediate cytotoxic activity via the production of ROS (Ito et al., 1992). Therefore, low levels of SiUCP2 expression in the coelomocytes, Aristotles' lantern and stomach may be related to ROS.
Lipids, including fat, phospholipids, and sterols, play a crucial role in individual life activities. The accumulation of total lipids, unsaturated FAs, and phospholipids in marine animals is essential for gonadal and early development (Palacios et al., 2007; Farhoudi et al., 2011). Previous studies have shown that the use of FAs in embryos varied with embryonic development stages (Yao and Zhao, 2006). During the early development of stingray embryos for example, high PUFA levels occur in the blastula stage, the gastrula stage and organogenesis, indicating that more PUFAs are needed during these periods to support the energy required for embryonic development (Yao et al., 2009). To understand the expression of the SiUCP2 gene and its role in the development of embryos in S. intermedius, the relative expression levels of SiUCP2 gene at different development stages of S. intermedius were determined using qRTPCR. The results show that SiUCP2 was expressed at all development stages in sea urchins. Decreased SiUCP2 expression in early embryonic development may be related to fat utilization as an energy source, as developing embryos use their lipid reserves to meet their energy needs (Coulibaly et al., 2006). During the gastrula stage in sea urchins, the intestine is differentiated and they begin to take in external nutrients and FAs are synthesized. Meanwhile, lipid metabolism is relatively enhanced, so SiUCP2 gene expression was slightly increased (Zuo et al., 2016). When sea urchins develop into prism larvae, they begin to float. When they develop into two-arm larvae, the range of the sea urchin movement is expanded, and the digestive tract is formed in the larvae, so their food intake is increased, thus lipid and energy metabolism are enhanced (Chang et al., 2004). Therefore, the expression level of SiUCP2 gene was increased. When sea urchins undergo metamorphosis to the juvenile period, they became benthic, and their food changes from planktonic single-celled algae to benthic diatoms. Their food intake tends to be stable, so does the lipid metabolism and balanced. Therefore, the expression level of the SiUCP2 gene was relatively decreased.
The intestines of sea urchins may be the site for FA synthesis (Han et al., 2019) and the gonads of sea urchins are the place where lipids are stored (Zuo et al., 2016). To understand clearly the role of SiUCP2 in lipid metabolism, the difference in expression of SiUCP2 in the intestines and gonads during starvation was analyzed using qRT-PCR. A previous study showed that UCP2 expression in the lungs and stomach of starved or polysaccharide-treated mice were increased (Pecqueur et al., 2001). In this study, as the starvation period was prolonged, SiUCP2 expression showed a pattern of first decreasing, then increasing, and decreasing again in the intestine and gonads. The sea urchins used food storage as energy after starvation of 0–7 d, and their metabolism was slowed down. Therefore, the expression level of SiUCP2 was decreased after starvation for 0–7 d. When starvation was extended to 7–14 d, the expression levels of SiUCP2 were gradually increased in the intestine and gonads, and the expression level in the gonads was greatly increased, suggesting that sea urchins may maintain their life activities by consuming lipids and proteins in the gonads and intestine after 7–14 d of starvation, which further indicated that SiUCP2 played a role in lipid metabolism (Solanes et al., 2003; Tang et al., 2013), and UCP2 reduced ROS production that mediated oxidative damage in mitochondria (Echtay et al., 2002). After 14–21 d, the nutrients in sea urchins are exhausted therefore SiUCP2 expression gradually decreases.
Song (2002) used in-situ hybridization to detect the expression of UCP2 in mouse liver. The results show that UCP2 was not expressed in mouse hepatocytes but expressed in macrophages under normal healthy conditions. When treated with lipopolysaccharides (LPS), UCP2 was expressed in mouse hepatocytes. In this study, DIG-labeled specific SiUCP2 probes were designed to perform fluorescence in-situ hybridization in the intestines and gonads of S. intermedius. The results show that SiUCP2 was expressed in the intestine and gonads, which is consistent with our qRT-PCR results. Owing to the scattered positive signals in the gonads, it was presumed that SiUCP2 was expressed in gonadal nutritive phagocytes, which is consistent with the findings of Larrouy et al. (1997). In the future, study of role of SiUCP2 in the gonads will be conducted.
In this study, the antibodies for SiUCP2 of S. intermedius were prepared successfully. Subsequently, we extracted the UCP2 protein and used Western blot to detect its expression in the intestines and gonads of S. intermedius. The results of SDS-PAGE show that the SiUCP2 protein with a molecular weight of 36.11 kDa was successfully obtained. This size was very similar to the molecular weight of human UCP2 (Pecqueur et al., 1999) and Chinese perch (Wen et al., 2002). Furthermore, the SiUCP2 protein was expressed in the gonads. The above-mentioned results are consistent with our in-situ hybridization results, which further demonstrated that UCP2 was closely related to lipid and energy metabolism.
Based on the results of this study, we speculated that the SiUCP2 gene might be related to the fatty acids synthesis of S. intermedius. In the future, we will continue to pay attention to SiUCP2, study which fatty acids are produced by SiUCP2, clarify the mechanism of SiUCP2 in the synthesis of fatty acids, improve the content of fatty acids in sea urchins, and further improve the gonadal quality of sea urchins. We hope to combine molecular technology with aquaculture in the future to select and breed more nutritious and economic value of S. intermedius.
5 DATA AVAILABILITY STATEMENTThe data used to support the findings of this study are available from the corresponding author upon request.
6 AUTHOR CONTRIBUTIONJun DING, Bing HAN, and Lingshu HAN contributed to the conception of the study. Lingshu HAN and Zijiao QUAN performed the experiments. Lingshu HAN, Zijiao QUAN, Xiaofang HUANG, and Beichen DING performed the data analyses. Lingshu HAN drafted the manuscript. Lingshu HAN, Jun DING, Bing HAN, and Heng WANG revised the manuscript. Jun DING and Yaqing CHANG approved the final version.
Andrews Z B, Liu Z W, Walllingford N, Erion D M, Borok E, Friedman J M, Tschöp M H, Shanabrough M, Cline G, Shulman G I, Coppola A, Gao X B, Horvath T L, Diano S. 2008. UCP2 mediates ghrelin's action on NPY/AgRP neurons by lowering free radicals. Nature, 454(7206): 846-851.
DOI:10.1038/nature07181 |
Brookes P S, Parker N, Buckingham J A, Vidal-Puig A, Halestrap A P, Gunter T E, Nicholls D G, Bernardi P, Lemasters J J, Brand M D. 2008. UCPs-unlikely calcium porters. Nature Cell Biology, 10(11): 1 235-1 237.
DOI:10.1038/ncb1108-1235 |
Campbell N A, Reece J B. 2005. Biology. 7th edn. New York: Pearson
|
Chang Y Q, Ding J, Song J, Yang W. 2004. Sea Cucumber, Sea Urchin Biology Research and Breeding. Beijing: Ocean Press (in Chinese)
|
Coppola A, Liu Z W, Andrews Z B, Paradis E, Roy M C, Friedman J M, Ricquier D, Richard D, Horvath T L, Gao X B, Diano S. 2007. A central thermogenic-like mechanism in feeding regulation: an interplay between arcuate nucleus T3 and UCP2. Cell Metabolism, 5(1): 21-33.
DOI:10.1016/j.cmet.2006.12.002 |
Coulibaly I, Gahr S A, Palti Y, Yao J B, Rexroad III C E. 2006. Genomic structure and expression of uncoupling protein 2 genes in rainbow trout (oncorhynchus mykiss). BMC Genomics, 7: 203.
DOI:10.1186/1471-2164-7-203 |
Douaire M, Le Fur N, El Khadir-Mounier C, Langlois P, Flamant F, Mallard J. 1992. Identifying genes involved in the variability of genetic fatness in the growing chicken. Poultry Science, 71(11): 1 911-1 920.
DOI:10.3382/ps.0711911 |
Echtay K S, Roussel D, St-Pierre J, Jekabsons M B, Cadenas S, Stuart J A, Harper J A, Roebuck S J, Morrison A, Pickering S, Clapham J C, Brand M D. 2002. Superoxide activates mitochondrial uncoupling proteins. Nature, 415(6867): 96-99.
DOI:10.1038/415096a |
Emre Y, Nübel T. 2010. Uncoupling protein UCP2: when mitochondrial activity meets immunity. FEBS Letters, 584(8): 1 437-1 442.
DOI:10.1016/j.febslet.2010.03.014 |
Farhoudi A, Kenari A M A, Nazari R M, Makhdoomi C H. 2011. Study of body composition, lipid and fatty acid profile during larval development in Caspian Sea Carp (Cyprinus carpio). Journal of Fisheries and Aquatic Science, 6(4): 417-428.
DOI:10.3923/jfas.2011.417.428 |
Fleury C, Sanchis D. 1999. The mitochondrial uncoupling protein-2: current status. The International Journal of Biochemistry & Cell Biology, 31(11): 1 261-1 278.
DOI:10.1016/s1357-2725(99)00049-7 |
Garlid K D, Jabrek M, Jezek P, Varecha M. 2000. How do uncoupling proteins uncouple?. BBA — Bioenergetics, 1459(2-3): 383-389.
DOI:10.1016/S0005-2728(00)00175-4 |
Geri G, Poli B M, Zappa A, Campodoni G, Franci O. 1990. Relationships between adipose tissue characteristics of newborn pigs and subsequent performance: III. Histological and chemical characteristics of backfat. Journal of Animal Science, 68(7): 1 936-1 943.
DOI:10.2527/1990.6871936x |
Han L S, Ding J, Wang H, Zuo R T, Quan Z J, Fan Z H, Liu Q D, Chang Y Q. 2019. Molecular characterization and expression of SiFad1 in the sea urchin (Strongylocentrotus intermedius). Gene, 705: 133-141.
DOI:10.1016/j.gene.2019.04.043 |
Huang Z G, Xie Z. 2004. Advances in research on genes related to fat traits in livestock and poultry. Animal Husbandry & Veterinary Medicine, 36(4): 41-43.
(in Chinese) |
Ito T, Matsutani T, Mori K, Nomura T. 1992. Phagocytosis and hydrogen peroxide production by phagocytes of the sea urchin Strongylocentrotus nudus. Developmental & Comparative Immunology, 16(4): 287-294.
DOI:10.1016/0145-305x(92)90003-u |
Jiang Z H, Michal J J, Tobey D J, Daniels T F, Rule D C, MacNeil M D. 2008. Significant associations of stearoyl-CoA desaturase (SCD1) gene with fat deposition and composition in skeletal muscle. International Journal of Biological Sciences, 4(6): 345-351.
DOI:10.7150/ijbs.4.345 |
Jiang Z Q, Jia Z M, Han Y B. 2002. The compensatory growth and its mechanismin of red drum, Sciaenops ocellatus, after food deprivation. Journal of Fisheries of China, 26(1): 67-72.
(in Chinese with English abstract) |
Kang D J, Zhou G X, Zhou S W, Zeng J, Wang X L, Jiang Y, Yang Y X, Chen Y L. 2017. Comparative transcriptome analysis reveals potentially novel roles of homeobox genes in adipose deposition in fat-tailed sheep. Scientific Reports, 7(1): 14 491.
DOI:10.1038/s41598-017-14967-9 |
Kelly M S, Hunter A J, Scholfield C L, McKenzie J D. 2000. Morphology and survivorship of larval Psammechinus miliaris (Gmelin) (Echinodermata: Echinoidea) in response to varying food quantity and quality. Aquaculture, 183(3-4): 223-240.
DOI:10.1016/s0044-8486(99)00296-3 |
Klingenberg M, Echtay K S. 2001. Uncoupling proteins: the issues from a biochemist point of view. Biochimica et Biophysica Acta (BBA)-Bioenergetics, 1504(1): 128-143.
DOI:10.1016/S0005-2728(00)00242-5 |
Lares M T, Pomory C M. 1998. Use of body components during starvation in Lytechinus variegatus, (Lamarck) (Echinodermata: Echinoidea). Journal of Experimental Marine Biology and Ecology, 225(1): 99-106.
DOI:10.1016/S0022-0981(97)00216-5 |
Larrouy D, Laharrague P, Carrera G, Viguerie-Bascands N, Levi-Meyrueis C, Fleury C, Pecqueur C, Nibbelink M, André M, Casteilla L, Ricquier D. 1997. Kupffer cells are a dominant site of uncoupling protein 2 expression in rat liver. Biochemical and Biophysical Research Communications, 235(3): 760-764.
DOI:10.1006/bbrc.1997.6852 |
Li H, Wu G, Zhang J, Yang N. 2010. Identification of the heart-type fatty acid-binding protein as a major gene for chicken fatty acid metabolism by Bayesian network analysis. Poultry Science, 89(9): 1 825-1 833.
DOI:10.3382/ps.2010-00699 |
Li X, Qin Y J, Li Y Y. 2004. Metabolism of sea urchin Strontgylocentrotus intermedius during starvation. Journal of Fishery Sciences of China, 11(4): 302-306.
(in Chinese with English abstract) |
Liang X F, Ogata H Y, Oku H, Chen J W, Hwang F. 2003. Abundant and constant expression of uncoupling protein 2 in the liver of red sea bream Pagrus major. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology, 136(3): 655-661.
DOI:10.1016/S1095-6433(03)00218-6 |
Liao W Q, Liang X F, Wang L, Ma X, Fang L, Li G S. 2006. cDNA sequence cloning and tissue expression of uncoupling protein 2 of silver carp (Hypophthalmichthys molitrix). Zoological Research, 27(4): 375-381.
(in Chinese with English abstract) |
Livak K J, Schmittgen T D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods, 25(4): 402-408.
DOI:10.1006/meth.2001.1262 |
Luévano-Martínez L A. 2012. Uncoupling proteins (UCP) in unicellular eukaryotes: true UCPs or UCP1-like acting proteins?. FEBS Letters, 586(7): 1 073-1 078.
DOI:10.1016/j.febslet.2012.03.009 |
Malavazos A E, Corsi M F, Ermetici F, Coman C, Sardanelli F, Rossi A, Morricone L, Ambrosi B. 2007. Proinflammatory cytokines and cardiac abnormalities in uncomplicated obesity: relationship with abdominal fat deposition. Nutrition, Metabolism and Cardiovascular Diseases, 17(4): 294-302.
DOI:10.1016/j.numecd.2006.01.001 |
Matsuda J, Hosoda K, Itoh H, Son C, Doi K, Tanaka T, Fukunaga Y, Inoue G, Nishimura H, Yoshimasa Y, Yamori Y, Nakao K. 1997. Cloning of rat uncoupling protein-3 and uncoupling protein-2 cDNAs: their gene expression in rats fed high-fat diet. FEBS Letters, 418(1-2): 200-204.
DOI:10.1016/S0014-5793(97)01381-1 |
McCGraham N. 1967. The metabolic rate of fasting sheep in relation to total and lean body weight, and the estimation of maintenance requirements. Australian Journal of Agricultural Research, 18(1): 127-136.
DOI:10.1071/AR9670127 |
Medvedev A V, Robidoux J, Bai X, Cao W H, Floering L M, Daniel K W, Collins S. 2002. Regulation of the uncoupling protein-2 gene in INS-1 beta-cells by oleic acid. The Journal of Biological Chemistry, 277(45): 42 639-42 644.
DOI:10.1074/jbc.M208645200 |
Mehner T, Wieser W. 1994. Energetics and metabolic correlates of starvation in juvenile perch (Perca fluviatilis). Journal of Fish Biology, 45(2): 325-333.
DOI:10.1111/j.1095-8649.1994.tb01311.x |
Palacios E, Racotta I S, Arjona O, Marty Y, Le Coz J R, Moal J, Samain J F. 2007. Lipid composition of the pacific lion-paw scallop, Nodipecten subnodosus, in relation to gametogenesis: 2.lipid classes and sterols. Aquaculture, 266(1-4): 266-273.
DOI:10.1016/j.aquaculture.2007.02.030 |
Pecqueur C, Alves-Guerra M C, Gelly C, Levi-Meyrueis C, Couplan E, Collins S, Ricquier D, Miroux B. 2001. Uncoupling protein 2, in vivo distribution, induction upon oxidative stress, and evidence for translational regulation. The Journal of Biological Chemistry, 276(12): 8 705-8 712.
DOI:10.1074/jbc.M006938200 |
Pecqueur C, Cassard-Doulcier A M, Raimbault S, Miroux B, Fleury C, Gelly C, Bouillaud F, Ricquier D. 1999. Functional organization of the human uncoupling protein-2 gene, and juxtaposition to the uncoupling protein-3 gene. Biochemical and Biophysical Research Communications, 255(1): 40-46.
DOI:10.1006/bbrc.1998.0146 |
Santos R, da Costa G, Franco C, Gomes-Alves P, Flammang P, Coelho A V. 2009. First insights into the biochemistry of tube foot adhesive from the sea urchin Paracentrotus lividus (Echinoidea, Echinodermata). Marine Biotechnology, 11(6): 686-698.
DOI:10.1007/s10126-009-9182-5 |
Solanes G, Pedraza N, Iglesias R, Giralt M, Villarroya F. 2003. Functional relationship between MyoD and peroxisome proliferator-activated receptor-dependent regulatory pathways in the control of the human uncoupling protein-3 gene transcription. Molecular Endocrinology, 17(10): 1 944-1 958.
DOI:10.1210/me.2002-0395 |
Song Y. 2002. The Effects of Lipopolysaccharide and Indomethacin to the Body Temperature and the UCP2mRNA Expression in the Livers of Mice. China Medical University, Shenyang. (in Chinese with English abstract)
|
Stuart J A, Harper J A, Brindle K M, Brand M D. 1999. Uncoupling protein 2 from carp and zebrafish, ectothermic vertebrates. Biochimica et Biophysica Acta (BBA)-Bioenergetics, 1413(1): 50-54.
DOI:10.1016/S0005-2728(99)00081-X |
Tagen M, Elorza A, Kempuraj D, Boucher W, Kepley C L, Shirihai O S, Theoharides T C. 2009. Mitochondrial uncoupling protein 2 inhibits mast cell activation and reduces histamine content. The Journal of Immunology, 183(10): 6 313-6 319.
DOI:10.4049/jimmunol.0803422 |
Tang Z G, Sun C Y, Yan A F, Wu S G, Qin C B, Zhang Y H, Li W S. 2013. Genes involved in fatty acid metabolism: molecular characterization and hypothalamic mRNA response to energy status and neuropeptide Y treatment in the orange-spotted grouper Epinephelus coioides. Molecular and Cellular Endocrinology, 376(1-2): 114-124.
DOI:10.1016/j.mce.2013.06.020 |
Taouis M, Chen J W, Daviaud C, Dupont J, Derouet M, Simon J. 1998. Cloning the chicken leptin gene. Gene, 208(2): 239-242.
DOI:10.1016/s0378-1119(97)00670-7 |
Thompson M P, Kim D. 2004. Links between fatty acids and expression of UCP2 and UCP3 mRNAs. FEBS Letters, 568(1).
DOI:10.1016/j.febslet.2004.05.011 |
Walker C W, Lesser M P, Unuma T. 2013. Sea urchin gametogenesis-structural, functional and molecular/genomic. Developments in Aquaculture and Fisheries Science, 38: 25-43.
DOI:10.1016/B978-0-12-396491-5.00003-4 |
Wang Y, Nishi M, Doi A, Shono T, Furukawa Y, Shimada T, Furuta H, Sasaki H, Nanjo K. 2010. Ghrelin inhibits insulin secretion through the AMPK-UCP2 pathway in beta cells. FEBS letters, 584(8).
DOI:10.1016/j.febslet.2010.02.069 |
Wen X B, Chen L Q, Ai C X, Zhou Z L. 2002. Starvation metabolism in parent Chinese mitten-handed crab (Eriocheir sinensis). Chinese Journal of Applied Ecology, 13(11): 1 441-1 444.
(in Chinese with English abstract) |
Werner P, Neuenschwander S, Stranzinger G. 1999. Characterization of the porcine uncoupling proteins 2 and 3 (UCP2 & UCP3) and their localization to chromosome 9 p by somatic cell hybrids. Animal Genetics, 30(3): 221-224.
DOI:10.1046/j.1365-2052.1999.00462.x |
Yao J J, Zhao Y L, Li C, He D J, Hu C Y. 2009. Changes in fatty acid composition during early embryonic development of yellow catfish Pelteobagrus fiulvidraco. Fisheries Science, 28(11): 644-647.
(in Chinese with English abstract) DOI:10.3969/j.issn.1003-1111.2009.11.007 |
Yao J J, Zhao Y L. 2006. Changes of lipid content and fatty acid composition during embryonic development of Macrobrachium rosenbergii. Journal of East China Normal University (Natural Science), (4): 103–109. (in Chinese with English abstract)
|
Zhang B, Sun Y, Tang Q S. 2000. The effects of starvation on growth and biochemical composition in Pagrosomus major. Journal of Fisheries of China, 24(3): 206-210.
(in Chinese with English abstract) |
Zhou W, Sun J, Wang J J, Du J Y. 2008. Current status and challenges of sea urchin culture in China. Fisheries Science, 27(3): 151-153.
(in Chinese) |
Zuo R T, Hou S Q, Chang Y Q, Ding J, Song J, Zhao C, Zhang W J. 2016. Research advance of nutritional physiology in sea urchins: a review. Journal of Dalian Ocean University, 31(4): 463-468.
(in Chinese with English abstract) DOI:10.16535/j.cnki.dlhyxb.2016.04.019 |
 2021, Vol. 39
2021, Vol. 39