Institute of Oceanology, Chinese Academy of Sciences
Article Information
- ZHANG Xiaoyong, LI Yiyang, YU Zonghe, LIANG Xiao, QI Shuhua
- Phylogenetic diversity and bioactivity of culturable deepsea-derived fungi from Okinawa Trough
- Journal of Oceanology and Limnology, 39(3): 892-902
- http://dx.doi.org/10.1007/s00343-020-0003-z
Article History
- Received Jan. 29, 2020
- accepted in principle Apr. 2, 2020
- accepted for publication May. 21, 2020
2 Key Laboratory of Tropical Marine Bio-resources and Ecology/RNAM Center for Marine Microbiology/Guangdong Key Laboratory of Marine Material Medical, South China sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou 510301, China;
3 Southern Marine Science and Engineering Guangdong Laboratory(Guangzhou), Guangzhou 511458, China
Despite the absence of sunlight irradiation, deepsea environments are now recognized as highly dynamic, harboring a variety of unique organisms (Huang et al., 2020). In particular, the discovery of hydrothermal vents in deep oceans has resulted in some new concepts for considering energy sources available for sustaining life (Nagano and Nagahama, 2012). Nowadays, it is very clear that the steep chemical and physical gradients at deep-sea hydrothermal vent sites support a remarkably diverse microbial community (Pettit, 2011). Raghukumar et al. (2010) reported the diverse deep-sea microorganisms could have a complex adaptive mechanism and play a very important ecological role in various deep-sea environments.
To adequately investigate the diversity and ecological functions of microbes associated with the extraordinary deep-sea environments, various culturedependent and culture-independent methods have been developed. Comparison with culture-dependent approaches, culture-independent methods can discover the entire microbial community, including that of yet-to-be cultured microbes (Hao et al., 2019; Zhang et al., 2020a). Recently, many studies reported that many novel microbial phylotypes including BCGI clade and DSF-group (Xu et al., 2014), Rozella, and KML11 clade (Lara et al., 2010) were found by cultured-independent methods. However, the culturedependent approaches remain as the method of choice to study the bioactive metabolic requirement and biochemical characteristics of isolated fungal strains associated with the extraordinary deep-sea environments.
Searching marine-derived microorganisms with various bioactivities is always one of important objectives of our ongoing investigations on marine natural products. Deep-sea-derived fungi attracted our attention since many deep-sea fungal isolates from the South China Sea (Zhang et al., 2013) and the East Indian Ocean (Zhang et al., 2014) exhibited relatively diverse bioactivities in our previous studies. The aim of the present study is to investigate the phylogenetic diversity and bioactivity of culturable deep-sea-derived fungi from Okinawa Trough. The Okinawa Trough is regarded as a back arc basin formed behind the Ryukyu Trench (Shao et al., 2015), and it contains several active hydrothermal fields, including those in Ieya North and Iheya Ridge (Zhang et al., 2015). Recently, several hydrothermal mounds with vents and diffusing flows in Okinawa Trough have been widely documented (Glasby and Notsu, 2003; Inagaki et al., 2004). Moreover, biodiversity of microbial communities mainly including bacteria and archaea have been investigated in the area (Nagahama et al., 2006; Yanagawa et al., 2014). However, the culturable deep-sea-derived fungal communities from Okinawa Trough and their bioactivities have been rarely reported. In this study, we investigated the phylogenetic diversity, distribution, and bioactivity (including anti-bacterial-growth, anti-bacterial biofilm forming and anti-larval-settlement) of culturable fungi in four deep-sea sediment samples from Okinawa Trough.
2 MATERIAL AND METHOD 2.1 Study sites and samplingFour deep-sea sediment samples from Okinawa Trough were collected in April, 2014. Among them, samples A and C were collected using electro hydraulic grab with underwater television camera, and samples B and D were collected using box sampler. The sampling sites and depths of samples A–D were 126.91°E, 27.81°N, ~1 190 m; 126.98°E, 27.80°N, ~1 330 m; 126.97°E, 27.55°N, ~1 387 m; 126.93°E, 27.57°N, ~1 589 m; respectively (Zhang et al., 2016). Only one sample for each sampling site was collected in the Okinawa Trough cruise due to the extreme environments or weather conditions. It is the limit of this study. However, the four samples have been successfully used to investigate the bacterial diversities (Zhang et al., 2015) and cultureindependent fungal diversity (Zhang et al., 2016) to explore more deep-sea-derived microbial resources.
Distances from Samples A and B to the active hydrothermal vents in Iheya North were about 1.1 km and 6.3 km, respectively. And the distances from Samples C and D to the active hydrothermal vents in Iheya Ridge were about 0.4 km and 4.7 km, respectively (Zhang et al., 2015). The concentrations of several metal elements, including Mn, As, and other elements, in these sediment samples were determined by Zhang et al. (2015).
2.2 Fungal isolationCultivable fungi in the sediment samples were isolated by the particle plating technique described by Cathrine and Raghukumar (2009). Fungi were incubated on four different isolation media, including mPDA, mCDA, mMEA, and mGPSA (Zhang et al., 2013), and each medium was used at 1/5 strength in order to simulate the oligotrophic condition in deepsea sediments. After incubated in the dark at 10 ℃ for 7–30 d, these different isolates were selected and transferred on new agar plates based on the differences in morphological traits of the fungal colonies, mainly including growth characteristics, diffusible pigments of spores and mycelia (Wang et al., 2011). In order to perform "dereplication", only one isolate was picked up when there were two or more isolates looked like the same taxon in morphological traits in a same agar plate. A more detailed method of fungal isolation was described in a previous study (Zhang et al., 2013).
2.3 Fungal identificationTo identify the selected fungal isolates, the fungal DNA were extracted, and the internal transcribed spacer (ITS) sequences were amplified by PCR and sequenced by traditional (Sanger) sequencing method (Danielsen et al., 2012). The total genomic DNA of each selected isolate was extracted from about 1.0 g of fungal mycelia according to the manufacturer's protocol (Pirttilä et al., 2001). After then, the fungal ITS sequence of each selected isolate was amplified with the universal primers ITS1 and ITS4 by PCR (Kim et al., 2013). A more detailed method of fungal ITS sequence amplification was described in a previous study (Zhang et al., 2013). The amplification product of ITS sequence from each selected isolate was sequenced by Invitrogen (China), and then the ITS sequence was analyzed and compared by BLASTs in GenBank to identify the selected fungal isolates at genus or species level (Song et al., 2019a).
After identified by ITS sequences, the selected fungal isolates were confirmed by morphological observation according to fungal morphological criteria by Wei (1979). Each selected fungal isolates was transferred onto a new mPDA plate (Zhang et al., 2013). After cultivated at 10 ℃ for 7–30 d, the morphological traits (visible examination) of the pure fungal colony, such as morphology, mycelia, and diffusible pigments, were examined by microscopy (Wang et al., 2011).
2.4 Phylogenetic and data analysisPhylogenetic analysis of the ITS sequences from 15 fungal representatives (15 different fungal species) isolated in this study was performed by software MEGA 5.0 (Tamura et al., 2011; Tejesvi et al., 2011). The dissimilarity of fungal community in samples A, B, C, and D were estimated by Bray-Curtis analysis using SPSS for Window Soft (Version 11.5) (ToledoHernández et al., 2008). Fungal ITS sequences of the 15 representative (15 different fungal species) isolates were deposited in GenBank under accession numbers KX258798–KX258812.
2.5 Fermentation and extractionTo obtain more diverse metabolites from the fungal isolates, two different media were applied for fermentation. Each selected fungal isolate was inoculated in 500-mL Erlenmeyer flasks containing 150 mL of Medium A (glucose 0.2%, maltose 0.4%, mannitol 0.4%, monosodium glutamate 0.2%, KH2PO4 0.05%, MgSO4·7H2O 0.003%, corn steep liquor 0.01%, yeast extract 0.06%, and sea salt 3%) or Medium B (glucose 0.4%, potato 4%, and sea salt 3%) (Nong et al., 2013), and then cultivated on a rotary shaker at 200 r/min and 15 ℃ for 10 d. About 150 mL of fungal culture broths were filtered to separate the fungal mycelia and broth supernatant. The broth supernatant and mycelia from each representative isolate were extracted with ethyl acetate and 80% acetone, respectively. More detailed information of extraction was described by Zhang et al. (2019). Thirty crude extracts were obtained from the 15 representative isolates cultured in Media A and B.
2.6 Determination of anti-bacterial-growth activityAs an initial screening method, the anti-bacterialgrowth activity of the 30 extracts were determined against a larval settlement-inducing bacterium Micrococcus luteus UST950701-006 (ML-006) (Qi et al., 2009) and a biofilm-forming bacterium Shewanella onedensis MR-1 (SO-MR-1) (Thormann et al., 2004) using the standard paper-disk-diffusion technique (D'Souza et al., 2010). After impregnated with 50 μg of tested extracts (in ethyl acetate), the disks (6 mm in diameter) were placed on the agar plate containing indicator bacteria. After incubated for 1 d at 30 ℃, the diameter of the bacterial growth inhibition zone was determined as the anti-bacterial-growth activity of the crude extracts. The test bacteria ML-006 and SO-MR-1were inoculated on PY (peptone 0.5%, yeast extract 0.3%, sea salt 3%) agar medium. Detailed method of detecting anti-bacterial-growth activity was described by Dash et al. (2009).
After determined the diameter of the bacterial growth inhibition zone, the extracts with strong antibacterial-growth activity were further evaluated for the minimum inhibitory concentrations (MICs) by the microtitre plate assay (Sarker et al., 2007; Zeng et al., 2008). Briefly, the indicator bacterium was cultivated in PY medium for 12 h (150 r/min, 30 ℃), and adjusted to an optical density 0.6 at 560 nm with PY medium as indicator bacterial suspension. About 1 μL of indicator bacterial suspension and 0.5 μL of selected active extracts (in dimethyl sulfoxide) were transferred to each well (consisting of 100-μL PY medium) of 96-well polystyrene microtitre plates (the final concentrations of the extracts were 50, 25, 12.5, 6.2, 3.1, and 1.6 μg/mL) and incubated at 30 ℃ for 24 h. Streptomycin sulfate was selected as the positive control. A more detailed method of MIC determination was described by Zhang et al. (2019).
2.7 Anti-bacterial biofilm formation assayAnti-bacterial biofilm formation activity of all the extracts was evaluated using the standard sterile 24-well polystyrene plates (Burmølle et al., 2006). The indicator bacterium for biofilm formation is SO-MR-1 (Thormann et al., 2004). Briefly, 149 μL of sterilized water, 1 μL of concentrated extract solution (10 mg/mL with dimethyl sulfoxide (DMSO)) and 50 μL of bacterial inoculums were added into one well of the sterile 24-well polystyrene plates (the final concentration of the tested extract was 50 μg/mL). After the allotted incubation period (at 30 ℃ for 2 d), the plate with the biofilm was rinsed with phosphate buffer saline (pH 7.2) and stained with crystal violet (Burmølle et al., 2006). The inhibiting effects of the extracts on biofilm forming were evaluated as the ratio of the absorbance (at 600 nm) of attached cells to that of planktonic cells (O'Toole and Kolter, 1998). More detailed information of anti-bacterial biofilm formation assay was described by Dash et al. (2009).
2.8 Anti-larval-settlement assayThe direct anti-larval-settlement activity of the fungal extracts was tested using the marine macrofoulers bryozoan Bugula neritina and barnacle Balanus amphitrite. Colonies of B. neritina and B. amphitrite were collected from Daya Bay (114.54°E, 22.67°N), Shenzhen, China. After exposed to sunlight for about 30 min, the larvae of B. neritina were released by adult colonies and collected to test (Bryan et al., 1998). The larvae of B. amphitrite was raised to competence according to Thiyagarajan et al. (2003).
The anti-larval-settlement activities of 30 extracts against B. neritina and B. amphitrite were evaluated using 24-well polystyrene plates (Zhang et al., 2019). Briefly, 999 μL of filtered sea water, 1 μL of concentrated extract solution (10 mg/mL with DMSO) and 20 competent larvae were transferred into each well of the sterile 24-well polystyrene plates (the final concentration of the tested extract was 50 μg/mL). After incubated at 25 ℃ for 1 h for B. neritina and 2 d for B. amphitrite, the anti-larval-settlement effects of each test extract was determined by examining the settled ratio of the larvae. More detailed information of anti-larval-settlement assay was described in a previous study by Zhang et al. (2019).
After screened for anti-larval settlement activity, these extracts with strong activity were further examined their EC50 and LC50 values of anti-larval settlement activity against B. neritina and B. amphitrite. All selected extracts were diluted to a series of concentrations (50, 25, 12.5, 6.2, 3.1, and 1.6 μg/mL) with filtered seawater and further determined their anti-larval settlement activity as described by a previous study (Zhang et al., 2019). The calculation of EC50 and LC50 of anti-larval settlement activity of each selected extract was described by Xu et al. (2010).
3 RESULT AND DISCUSSION 3.1 Fungal isolation and phylogenetic diversity in deep-sea sedimentsSeventy-six fungal isolates were recovered from the four different deep-sea sediments from Okinawa Trough. All the 76 isolates were identified by ITS sequencing, and the ITS sequences of the 76 isolates were BLAST searched in GenBank. The results showed that almost all of isolates shared 99%–100% sequence similarities with known fungal strains in GenBank. After confirmed by fungal morphological observation (data not shown), the 76 identified isolates were assigned to 15 fungal taxa of six fungal genera including Aspergillus, Cladosporium, Mycosphaerella, Penicillium, Purpureocillium, and Schizophyllum (Table 1 and Fig. 1), suggesting there were abundant and diverse fungi in deep-sea sediments from Okinawa Trough. There was some evidence that many marine fungal species could be accumulated in deep-sea sediments (Shang et al., 2012). In a previous study, Damare and Raghukumar (2008) reported that fungal biomass carbon in deepsea sediments could be evaluated by detecting and calculating the length and width of fungal mycelia using a calibrated ocular micrometer, which provided a directed evidence that fungi could be abundantly found in deep-sea sediments and played a very important ecological role in aggregate formation of deep-sea sediments.
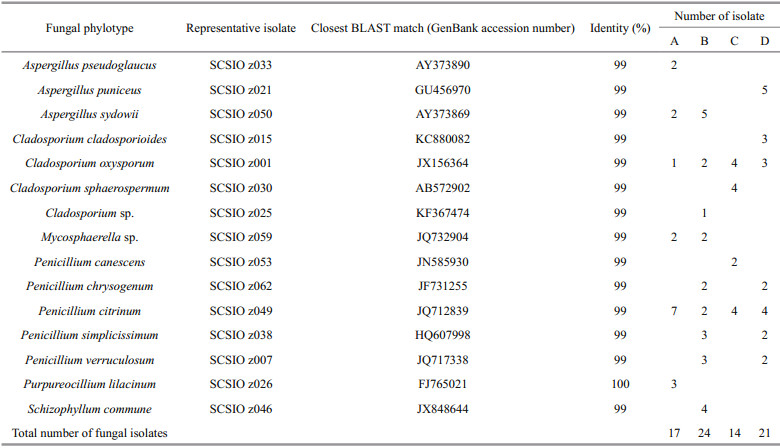
|

|
| Fig.1 Neighbor-joining phylogenetic tree from analysis of the ITS sequences of culturable fungi isolated from deep-sea sediments from Okinawa Trough The numbers at nodes are percentages indicating the levels of bootstrap support, based on a neighbor joining analysis of 1 000 resampled datasets. Only values of >50% are shown. Scale bar: 0.05 substitutions per nucleotide position. |
Among the six genera, Penicillium, Cladosporium, and Aspergillus were the most abundant and diverse fungal genera in the four different deep-sea sediments from Okinawa Trough. Many species belonged to the three genera are now considered widespread in marine environments (Pang et al., 2016), such as coral reefs (Zhang et al., 2019), mangrove ecosystem (Vanegas et al., 2019) and deep-sea sediments (Zhang et al., 2016). The remaining three genera, including Mycosphaerella, Purpureocillium and Schizophyllum, were relatively rare in the deep-sea sediments from Okinawa Trough. Mycosphaerella frequently found in soils or plants is known as one of the largest genera of plant pathogen fungi (Zhan et al., 2003). However, it is a new record for deep-sea-derived fungi in this study. Purpureocillium were frequently found in marine corals, sponges, and plants from shallow seawater (Yu et al., 2013). As marine original fungi, Schizophyllum spp. were frequently found in sea grass and algae (Poli et al., 2018), and might play a critical role in degrading hemicelluloses. All the fungal taxa recovered in this study were belonged to phylum Ascomycota and Basidiomycota (Fig. 1), indicating the both fungal phyla were the dominant fungal community in the deep-sea sediments from Okinawa Trough. This result was in agreement with previous reports on fungal diversity of deep-sea environments from Lau Basin (Burgaud et al., 2009), East Pacific Rise, and Mid-Atlantic Ridge (Le Calvez et al., 2009).
The fungal taxa isolated by culture-dependent in this study approach were much less than those were recovered by a high-throughput Illumina sequencing in an our previous study, which revealed that about 176 fungal taxa were recovered from the sediments (Zhang et al., 2016). The main reasons might be that most of deep-sea-derived fungi were hardly isolated and cultured using a culture-dependent approach. By further comparison with fungal community recovered by high-throughput Illumina sequencing (Zhang et al., 2016), several fungal taxa isolated in this study, such as Mycosphaerella sp. and Penicillium lilacinum, were not recovered by high-throughput Illumina sequencing, suggesting the greater fungal diversity could be recovered by combining the high-throughput sequencing technology with a traditional culturedependent approach.
3.2 Distribution and comparison of fungal communityMost fungal taxa were distributed in one or two sediment samples, while C. oxysporum and Penicillium citrinum could were recovered from all the four sediment samples (Table 1), indicating the two fungal taxa were widely distributed in the deepsea sediments from Okinawa Trough. The Bray-Curtis analysis showed 44.3%–89.2% dissimilarity of fungal communities (Table 2), indicating the diversities and distributions of fungal communities varied according to the different sites. As an effective analysis method for multivariate ecological data (Beals, 1984), BrayCurtis dissimilarity could widely applied in the comparison of microbial community in sediments (Liu et al., 2018), fish eggs and larvae in the Huanghe (Yellow) River estuary, China (Song et al., 2019b) and intertidal macrobenthos in the Naf River estuary, Bangladesh (Noman et al., 2019).
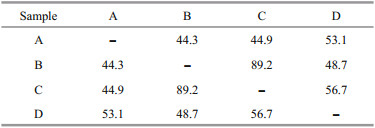
|
Further comparison analysis showed the numbers of fungal isolates and taxa recovered from samples B and D were relatively higher than that from Samples A and C (Table 1), suggesting that the distance from hydrothermal vents might play an important role in restructuring fungal community in deep sea sediments. Furthermore, the numbers of species and isolates of genus Pencillium (recognized as Mn-oxidizing fungi) in Samples B and D were relatively higher than that in Samples A and C. In a previous study, the concentration of Mn in Samples B and D was obviously higher than that in Samples A and C (Zhang et al., 2015). Our results in this study suggest that the metal elements might play an important role in constructing the fungal community in the deep-sea sediments.
3.3 Bioactivity of deep-sea-derived fungi from Okinawa TroughRecently, deep-sea-derived fungi have gradually become an important resource for bioactive compounds (Chen et al., 2020; Han et al., 2020; Pang et al., 2020). However, systematic bioactive investigations were rarely reported. In this study, 30 extracts of 15 fungal representative isolates from deep-sea sediments in Okinawa Trough were screened for anti-bacterial-growth, anti-bacterial biofilm forming and anti-larval-settlement activities. About 75% of the extracts displayed distinct bioactivity, emphasizing the potentials of the deep-sea fungi from Okinawa Trough as producers of bioactive metabolites (Table 3).
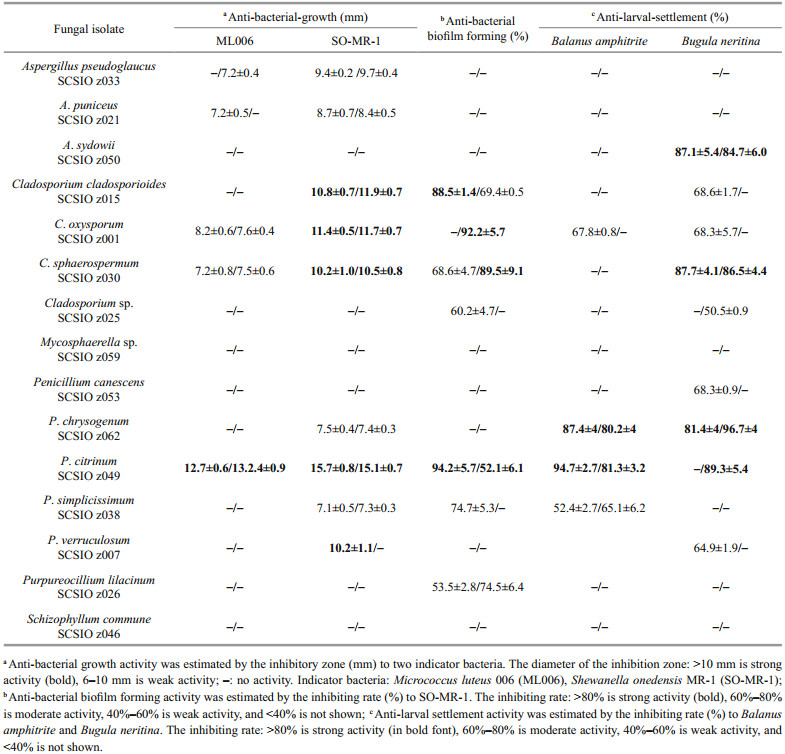
|
Notably, P. citrinum SCSIO z049 displayed a strong and wide range of inhibit activity against all tested indicator bacteria and marine macrofoulers (Tables 3 & 4). In addition, Penicillium verruculosum SCSIO z007 exhibited a strong anti-bacterial-growth activity against SO-MR-1 with the MIC value of 15.6 μg/mL, while Penicillium chrysogenum SCSIO z062 displayed a strong anti-larval-settlement activity against B. amphitrite and B. neritina with the EC50 value of 11.6–12.8 μg/mL (Table 4). These results suggest that deep-sea-derived Penicillium spp. from Okinawa Trough might produce a large number of various compounds with a wide range of biological activities. In recent studies, many novel compounds with anti-bacterial activity were isolated from the metabolites produced by Penicillium strains (Hou et al., 2019; Zhang et al., 2020b). For examples, two novel compounds penicilone H and penicipyrrodiether A, isolated from Penicillium janthinellum HK1-6 and Penicillium sp. ZZ380, displayed a very strong antibacterial activity against several Grampositive indicator bacteria (Song et al., 2018; Chen et al., 2019). Known as an antimicrobial extrolite producer, P. verruculosum had 11 447 predicted protein-coding genes, of which 225 were polysaccharide degrading enzymes (Hu et al., 2016).
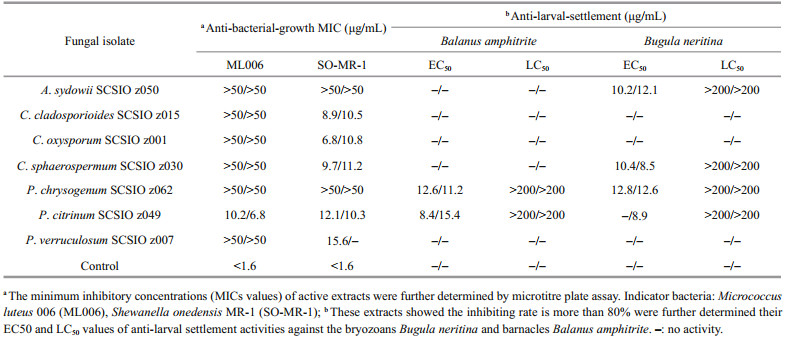
|
In addition, Penicillium, Cladosporium isolates recovered in this study also exhibited a wide spectrum of bioactivities. Especially, C. oxysporum SCSIO z001 displayed distinct inhibit activity against all tested indicator bacteria and marine macrofoulers (Tables 3 & 4). In addition, C. cladosporioides SCSIO z015 and C. sphaerospermum SCSIO z030 exhibited a strong anti-bacterial-growth activity with a MIC value of 8.9–11.2 μg/mL against SO-MR-1 (Table 4). Meanwhile, C. cladosporioides SCSIO z015 displayed a strong anti-bacterial biofilm forming activity against SO-MR-1. Recently, an increasing number of novel active compounds were found in the metabolites from marine-derived Cladosporium isolates. For examples, Li et al. (2017) isolated four novel cladosporol derivatives, cladosporols F–I from marine fungus C. cladosporioides EN-399, and found the four compounds displayed distinct antibacterial activity against three indicator bacteria. It was reported that a marine fungus Cladosporium sp. F14 could produce various kinds of compounds with antibacterial biofilm forming activity (Qi et al., 2009).
Literature surveys showed that genus Aspergillus was characterized by its worldwide and frequent distribution, and its species were known to generate a very wide spectrum of bioactivities, such as antibacterial-growth (Li et al., 2019; Song et al., 2019c), anti-bacterial biofilm forming acticity (Loges et al., 2020), and antifouling activity (Nong et al., 2015). However, three Aspergillus isolates exhibited only anti-bacterial-growth or anti-larval-settlement activities in this study (Table 3). One of the main reasons might be that different cultivation methods, including media compositions, culture vessel, and culture time, could affect fungal strains' producing secondary metabolites. Recently, one strain many compounds (OSMAC) approach was widely applied in finding diverse microbial metabolites (Hewage et al., 2014; Romano et al., 2018). Bovio et al. (2019) systematic investigated the sponge fungal biodiversity and chemodiversity by OSMAC approach and liquid chromatography-tandem mass spectrometry (LC-MS/MS), and found the sponge-associated fungal isolate fungus Eurotium chevalieri MUT 2316 displayed an astonishing metabolic diversity and promising bioactivity against some Gram-positive bacteria.
4 CONCLUSIONSeventy-six isolates belonging to 15 fungal taxa were revealed from a relatively abundant and diverse fungal community in the deep-sea sediments form Okinawa Trough. Aspergillus, Cladosporium, and Penicillium were the dominant fungal genera, while Mycosphaerella, Purpureocillium, and Schizophyllum were relatively rare in the deep-sea sediments from Okinawa Trough. Moreover, about 75% of the extracts from the 15 fungal representative isolates displayed distinct bioactivities, including anti-bacterial-growth, anti-bacterial biofilm forming and anti-larvalsettlement activities. Cladosporium and Penicillium isolates displayed a relatively strong and wide spectrum of bioactivities against all the test indicator bacteria and marine macrofoulers, while other fungal isolates exhibited only anti-bacterial-growth or antilarval-settlement activities.
5 DATA AVAILABILITY STATEMENTThe datasets that are not publicly available during the current study can be available from the corresponding author on reasonable request.
6 ACKNOWLEDGMENTThe authors are grateful to the R/V Kexue of the Chinese Academy of Sciences for collecting samples and WPOS sample center for providing samples.
Beals E W. 1984. Bray-Curtis ordination: an effective strategy for analysis of multivariate ecological data. Adv. Ecol. Res., 14: 1-55.
|
Bovio E, Garzoli L, Poli A, Luganini A, Villa P, Musumeci R, McCormack G P, Cocuzza C E, Gribaudo G, Mehiri M, Varese G C. 2019. Marine fungi from the sponge Grantia compressa: biodiversity, chemodiversity, and biotechnological potential. Mar. Drugs, 17(4): 220.
DOI:10.3390/md17040220 |
Bryan P J, Kreider J L, Qian P Y. 1998. Settlement of the serpulid polychaete Hydroides elegans (Haswell) on the arborescent bryozoan Bugula neritina (L.): evidence of a chemically mediated relationship. J. Exp. Mar. Biol. Ecol., 220(2): 171-190.
DOI:10.1016/S0022-0981(97)00105-6 |
Burgaud G, Le Calvez T, Arzur D, Vandenkoornhuyse P, Barbier G. 2009. Diversity of culturable marine filamentous fungi from deep-sea hydrothermal vents. Environ. Microbiol., 11(6): 1 588-1 600.
DOI:10.1111/j.1462-2920.2009.01886.x |
Burmølle M, Webb J S, Rao D, Hansen L H, Serensen S J, Kjelleberg S. 2006. Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms. Appl. Environ. Microbiol., 72(6): 3 916-3 923.
DOI:10.1128/AEM.03022-05 |
Cathrine S J, Raghukumar C. 2009. Anaerobic denitrification in fungi from the coastal marine sediments off Goa, India. Mycol. Res., 113(1): 100-109.
DOI:10.1016/j.mycres.2008.08.009 |
Chen M, Zheng Y Y, Chen Z Q, Shen N X, Shen L, Zhang F M, Zhou X J, Wang C Y. 2019. NaBr-induced production of brominated azaphilones and related tricyclic polyketides by the marine-derived fungus Penicillium janthinellum HK1-6. J. Nat. Prod., 82(2): 368-374.
DOI:10.1021/acs.jnatprod.8b00930 |
Chen W H, Liu H Y, Long J Y, Tao H M, Lin X P, Liao S R, Yang B, Zhou X F, Liu Y H, Wang J F. 2020. Asperpentenone A, A novel polyketide isolated from the deep-sea derived fungus Aspergillus sp. SCSIO 41024. Phytochem. Lett., 35: 99-102.
DOI:10.1016/j.phytol.2019.11.009 |
Damare S, Raghukumar C. 2008. Fungi and macroaggregation in deep-sea sediments. Microb. Ecol., 56: 168-177.
DOI:10.1007/s00248-007-9334-y |
Danielsen L, Thürmer A, Meinick P, Buée M, Morin E, Martin F, Pilate G, Daniel R, Polle A, Reich M. 2012. Fungal soil communities in a young transgenic poplar plantation form a rich reservoir for fungal root communities. Ecol. Evol., 2(8): 1 935-1 948.
DOI:10.1002/ece3.305 |
Dash S, Jin C L, Lee O O, Xu Y, Qian P Y. 2009. Antibacterial and antilarval-settlement potential and metabolite profiles of novel sponge-associated marine bacteria. J. Ind. Microbiol. Biotechnol., 36(8): 1 047-1 056.
DOI:10.1007/s10295-009-0588-x |
D'Souza F, Bruin A, Biersteker R, Donnelly G, Klijnstra J, Rentrop C, Willemsen P. 2010. Bacterial assay for the rapid assessment of antifouling and fouling release properties of coatings and materials. J. Ind. Microbiol. Biotechnol., 37(4): 363-370.
DOI:10.1007/s10295-009-0681-1 |
Glasby G P, Notsu K. 2003. Submarine hydrothermal mineralization in the Okinawa Trough, SW of Japan: an overview. Ore Geol. Rev., 23(3-4): 299-339.
DOI:10.1016/j.oregeorev.2003.07.001 |
Han W R, Cai J, Zhong W M, Xu G M, Wang F Z, Tian X P, Zhou X J, Liu Q C, Liu Y H, Wang J F. 2020. Protein tyrosine phosphatase 1B (PTP1B) inhibitors from the deep-sea fungus Penicillium chrysogenum SCSIO 07007. Bioorg. Chem., 96: 103646.
DOI:10.1016/j.bioorg.2020.103646 |
Hao W J, Wichels A, Fuchs B, Tang X X, Gerdts G. 2019. Bacterial community succession in response to dissolved organic matter released from live jellyfish. J. Oceanol. Limnol., 37(4): 1 229-1 244.
DOI:10.1007/s00343-019-8106-0 |
Hewage R T, Aree T, Mahidol C, Ruchirawat S, Kittakoop P. 2014. One strain-many compounds (OSMAC) method for production of polyketides, azaphilones, and an isochromanone using the endophytic fungus Dothideomycete sp. Phytochemistry, 108: 87-94.
DOI:10.1016/j.phytochem.2014.09.013 |
Hou X M, Li Y Y, Shi W W, Fang Y W, Chao R, Gu Y C, Wang C Y, Shao C L. 2019. Integrating molecular networking and 1H NMR to target the isolation of chrysogeamides from a library of marine-derived Penicillium Fungi. J. Org. Chem., 84(3): 1 228-1 237.
DOI:10.1021/acs.joc.8b02614 |
Hu L W, Taujale R, Liu F, Song J Z, Yin Q S, Zhang Y L, Guo J H, Yin Y B. 2016. Draft genome sequence of Talaromyces verruculosus ("Penicillium verruculosum") strain TS63-9, a fungus with great potential for industrial production of polysaccharide-degrading enzymes. J. Biotechnol., 219: 5-6.
DOI:10.1016/j.jbiotec.2015.12.017 |
Huang Y H, Sun C J, Yang G P, Yue X A, Jiang F H, Cao W, Yin X F, Guo C N, Niu J H, Ding H B. 2020. Geochemical characteristics of hadal sediment in the northern Yap Trench. J. Oceanol. Limnol., 38(3): 650-664.
DOI:10.1007/s00343-019-9010-3 |
Inagaki F, Takai K, Nealson K H, Horikoshi K. 2004. Sulfurovum lithotrophicum gen. nov., sp. nov., a novel sulfur-oxidizing chemolithoautotroph within the epsilon-Proteobacteria isolated from Okinawa Trough hydrothermal sediments. Int. J. Syst. Evol. Microbiol., 54(5): 1 477-1 482.
DOI:10.1099/ijs.0.03042-0 |
Kim D H, Kim S H, Kwon S W, Lee J K, Hong S B. 2013. Fungal diversity of rice straw for Meju fermentation. J. Microbiol. Biotechnol., 23(12): 1 654-1 663.
DOI:10.4014/jmb.1307.07071 |
Lara E, Moreira D, López-García P. 2010. The environmental Clade LKM11 and Rozella form the deepest branching clade of Fungi. Protist, 161(1): 116-121.
DOI:10.1016/j.protis.2009.06.005 |
Le Calvez T, Burgaud G, Mahé S, Barbier G, Vandenkoornhuyse P. 2009. Fungal diversity in deep-sea hydrothermal ecosystems. Appl. Environ. Microbiol., 75(20): 6 415-6 421.
DOI:10.1128/AEM.00653-09 |
Li H L, Li X M, Mándi A, Antus S, Li X, Zhang P, Liu Y, Kurtán T, Wang B G. 2017. Characterization of Cladosporols from the marine algal-derived endophytic fungus Cladosporium cladosporioides EN-399 and configurational revision of the previously reported cladosporol derivatives. J. Org. Chem., 82(19): 9 946-9 954.
DOI:10.1021/acs.joc.7b01277 |
Li J L, Jiang X, Liu X P, He C W, Di Y X, Lu S J, Huang H L, Lin B, Wang D, Fan B Y. 2019. Antibacterial anthraquinone dimers from marine derived fungus Aspergillus sp. Fitoterapia, 133: 1-4.
DOI:10.1016/j.fitote.2018.11.015 |
Liu H M, Wang B, Hu X K. 2018. Sediment bacterial communities are more complex in coastal shallow straits than in oceanic deep straits. J. Ocean. Limnol., 36(5): 1 643-1 654.
DOI:10.1007/s00343-018-7133-6 |
Loges L A, Silva D B, Paulino G V B, Landell M F, Macedo A J. 2020. Polyketides from marine-derived Aspergillus welwitschiae inhibit Staphylococcus aureus virulence factors and potentiate vancomycin antibacterial activity in vivo. Microb. Pathog., 143: 104066.
DOI:10.1016/j.micpath.2020.104066 |
Nagahama T, Hamamoto M, Horikoshi K. 2006. Rhodotorula pacifica sp. nov., a novel yeast species from sediment collected on the deep-sea floor of the north-west Pacific Ocean. Int. J. Syst. Evol. Microbiol., 56(1): 295-299.
DOI:10.1099/ijs.0.63584-0 |
Nagano Y, Nagahama T. 2012. Fungal diversity in deep-sea extreme environments. Fungal Ecol., 5(4): 463-471.
DOI:10.1016/j.funeco.2012.01.004 |
Noman M A, Mamunur R, Islam M S, Hossain M B. 2019. Spatial and seasonal distribution of intertidal macrobenthos with their biomass and functional feeding guilds in the Naf River estuary, Bangladesh. J. Ocean. Limnol., 37(3): 1 010-1 023.
DOI:10.1007/s00343-019-8063-7 |
Nong X H, Zhang X Y, Xu X Y, Qi S H. 2015. Antifouling compounds from the marine-derived fungus Aspergillus terreus SCSGAF0162. Nat. Prod. Commun., 10(6): 1 033-1 034.
|
Nong X H, Zheng Z H, Zhang X Y, Lu X H, Qi S H. 2013. Polyketides from a marine-derived fungus Xylariaceae sp. Mar. Drugs, 11(5): 1 718-1 727.
DOI:10.3390/md11051718 |
O'Toole G A, Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol., 28(3): 449-461.
DOI:10.1046/j.1365-2958.1998.00797.x |
Pang K L, Overy D P, Jones E B G, da Luz Calado M, Burgaud G, Walker A K, Johnson J A, Kerr R G, Cha H J, Bills G F. 2016. 'Marine fungi' and 'marine-derived fungi' in natural product chemistry research: toward a new consensual definition. Fungal Biol. Rev., 30(4): 163-175.
DOI:10.1016/j.fbr.2016.08.001 |
Pang X Y, Lin X P, Zhou X F, Yang B, Tian X P, Wang J F, Xu S H, Liu Y H. 2020. New quinoline alkaloid and bisabolane-type sesquiterpenoid derivatives from the deep-sea-derived fungus Aspergillus sp. SCSIO06786. Fitoterapia, 140: 104406.
DOI:10.1016/j.fitote.2019.104406 |
Pettit R K. 2011. Culturability and secondary metabolite diversity of extreme microbes: expanding contribution of deep sea and deep-sea vent microbes to natural product discovery. Mar. Biotechnol., 13(1): 1-11.
DOI:10.1007/s10126-010-9294-y |
Pirttilä A M, Hirsikorpi M, Kämäräinen T, Jaakola L, Hohtola A. 2001. DNA isolation methods for medicinal and aromatic plants. Plant Mol. Biol. Rep., 19(3): 273.
DOI:10.1007/BF02772901 |
Poli A, Vizzini A, Prigione V, Varese G S. 2018. Basidiomycota isolated from the Mediterranean Sea—phylogeny and putative ecological roles. Fungal Ecol., 36: 51-62.
DOI:10.1016/j.funeco.2018.09.002 |
Qi S H, Xu Y, Xiong H R, Qian P Y, Zhang S. 2009. Antifouling and antibacterial compounds from a marine fungus Cladosporium sp. F14. World J. Microbiol. Biotechnol., 25(3): 399-406.
DOI:10.1007/s11274-008-9904-2 |
Raghukumar C, Damare S R, Singh P. 2010. A review on deep-sea fungi: occurrence, diversity and adaptations. Bot. Mar., 53(6): 479-492.
|
Romano S, Jackson S A, Patry S, Dobson AD W. 2018. Extending the "one strain many compounds" (OSMAC) principle to marine microorganisms. Mar. Drugs, 16(7): 244.
DOI:10.3390/md16070244 |
Sarker S D, Nahar L, Kumarasamy Y. 2007. Microtitre plate-based antibacterial assay incorporating resazurin as an indicator of cell growth, and its application in the in vitro antibacterial screening of phytochemicals. Methods, 42(4): 321-324.
DOI:10.1016/j.ymeth.2007.01.006 |
Shang Z, Li X M, Meng L M, Li C S, Gao S S, Huang C G, Wang B G. 2012. Chemical profile of the secondary metabolites produced by a deep-sea sediment-derived fungus Penicillium commune SD-118. Chin. J. Ocean. Limnol., 30(2): 305-314.
DOI:10.1007/s00343-012-1075-1 |
Shao H B, Yang S Y, Wang Q, Guo Y L. 2015. Discriminating hydrothermal and terrigenous clays in the Okinawa Trough, East China Sea: evidences from mineralogy and geochemistry. Chem. Geol., 398: 85-96.
DOI:10.1016/j.chemgeo.2015.02.001 |
Song Q, Li X M, Hu X Y, Li X, Chi L P, Li H L, Wang B G. 2019c. Antibacterial metabolites from Ascidian-derived fungus Aspergillus clavatus AS-107. Phytochem. Lett., 34: 30-34.
DOI:10.1016/j.phytol.2019.09.004 |
Song T F, Chen M X, Ge Z W, Chai W Y, Li X C, Zhang Z Z, Lian X Y. 2018. Bioactive penicipyrrodiether A, an adduct of GKK1032 analogue and phenol A derivative, from a marine-sourced fungus Penicillium sp. ZZ380. J. Org. Chem., 83(21): 13 395-13 401.
DOI:10.1021/acs.joc.8b02172 |
Song Y F, Zhang L J, Luo X X. 2019b. Spatiotemporal distribution of fish eggs and larvae in the Huanghe (Yellow) River estuary, China in 2005-2016. J. Ocean. Limnol., 37(5): 1 625-1 637.
DOI:10.1007/s00343-019-8167-0 |
Song Y P, Miao F P, Liu X H, Ji N Y. 2019a. Responses of marine-derived Trichoderma fungi to seawater and their potential antagonistic behaviour. J. Ocean. Limnol., 37(2): 525-534.
DOI:10.1007/s00343-019-8059-3 |
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol., 28(10): 2 731-2 739.
DOI:10.1093/molbev/msr121 |
Tejesvi M V, Kajula M, Mattila S, Pirttilä A M. 2011. Bioactivity and genetic diversity of endophytic fungi in Rhododendron tomentosum Harmaja. Fungal Divers., 47: 97-107.
DOI:10.1007/s13225-010-0087-4 |
Thiyagarajan V, Harder T, Qiu J W, Qian P Y. 2003. Energy content at metamorphosis and growth rate of the early juvenile barnacle Balanus amphitrite. Mar. Biol., 143(3): 543-554.
DOI:10.1007/s00227-003-1077-9 |
Thormann K M, Saville R M, Shukla S, Pelletier D A, Spormann A M. 2004. Initial phases of biofilm formation in Shewanella oneidensis MR-1. J. Bacteriol., 186(23): 8 096-8 104.
DOI:10.1128/JB.186.23.8096-8104.2004 |
Toledo-Hernández C, Zuluaga-Montero A, Bones-González A, Rodríguez J A, Sabat A M, Bayman P. 2008. Fungi in healthy and diseased sea fans (Gorgonia ventalina): is Aspergillus sydowii always the pathogen?. Coral Reefs, 27(3): 707-714.
DOI:10.1007/s00338-008-0387-2 |
Vanegas J, Muñoz-García A, Pérez-Parra K A, Figueroa-Galvis I, Mestanza O, Polanía J. 2019. Effect of salinity on fungal diversity in the rhizosphere of the halophyte Avicennia germinans from a semi-arid mangrove. Fungal Ecol., 42: 100855.
DOI:10.1016/j.funeco.2019.07.009 |
Wang Y N, Shao C L, Zheng C J, Chen Y Y, Wang C Y. 2011. Diversity and antibacterial activities of fungi derived from the gorgonian Echinogorgia rebekka from the South China Sea. Mar. Drugs, 9(8): 1 379-1 390.
DOI:10.3390/md9081379 |
Wei J C. 1979. Fungi identification manual. Shanghai Science and Technology Press, Shanghai, China. 787p.
(in Chinese)
|
Xu W, Pang K L, Luo Z H. 2014. High fungal diversity and abundance recovered in the deep-sea sediments of the Pacific Ocean. Microb. Ecol., 68(4): 688-698.
DOI:10.1007/s00248-014-0448-8 |
Xu Y, He H P, Schulz S, Liu X, Fusetani N, Xiong H R, Xiao X, Qian P Y. 2010. Potent antifouling compounds produced by marine Streptomyces. Bioresour. Technol., 101(4): 1 331-1 336.
DOI:10.1016/j.biortech.2009.09.046 |
Yanagawa K, Breuker A, Schippers A, Nishizawa M, Ijiri A, Hirai M, Takaki Y, Sunamura M, Urabe T, Nunoura T, Takai K. 2014. Microbial community stratification controlled by the subseafloor fluid flow and geothermal gradient at the Iheya North Hydrothermal field in the Mid-Okinawa Trough (Integrated Ocean Drilling Program Expedition 331). Appl. Environ. Microbiol., 80(9): 6 126-6 135.
|
Yu Z S, Zhang B H, Sun W, Zhang F L, Li Z Y. 2013. Phylogenetically diverse endozoic fungi in the South China Sea sponges and their potential in synthesizing bioactive natural products suggested by PKS gene and cytotoxic activity analysis. Fungal Divers., 58: 127-141.
DOI:10.1007/s13225-012-0192-7 |
Zeng M Y, Cui W X, Zhao Y H, Liu Z Y, Dong S Y, Guo Y. 2008. Antiviral active peptide from oyster. Chin. J. Ocean. Limnol., 26(3): 307-312.
DOI:10.1007/s00343-008-0307-x |
Zhan J, Pettway R E, McDonald B A. 2003. The global genetic structure of the wheat pathogen Mycosphaerella graminicola is characterized by high nuclear diversity, low mitochondrial diversity, regular recombination, and gene flow. Fungal Genet. Biol., 38(3): 286-297.
DOI:10.1016/S1087-1845(02)00538-8 |
Zhang C, Zhang W J, Yin Q J, Li X G, Qi X Q, Wu L F. 2020a. Distinct influence of trimethylamine N-oxide and high hydrostatic pressure on community structure and culturable deep-sea bacteria. J. Oceanol. Limnol., 38(2): 364-377.
DOI:10.1007/s00343-019-9076-y |
Zhang J, Sun Q L, Zeng Z G, Chen S, Sun L. 2015. Microbial diversity in the deep-sea sediments of Iheya North and Iheya Ridge, Okinawa Trough. Microbiol. Res., 177: 43-52.
DOI:10.1016/j.micres.2015.05.006 |
Zhang P, Wei Q, Yuan X L, Xu K. 2020b. Reported alkaloids produced by marine-derived Penicillium species (covering 2014-2018). Bioorg. Chem, 99: 103840.
DOI:10.1016/j.bioorg.2020.103840 |
Zhang X Y, Hao H L, Lau S C K, Wang H Y, Han Y, Dong L M, Huang R M. 2019. Biodiversity and antifouling activity of fungi associated with two soft corals from the South China Sea. Arch. Microbiol., 201(6): 757-767.
DOI:10.1007/s00203-019-01639-7 |
Zhang X Y, Tang G L, Xu X Y, Nong X H, Qi S H. 2014. Insights into deep-sea sediment fungal communities from the East Indian Ocean using targeted environmental sequencing combined with traditional cultivation. PLoS One, 9(10): e109118.
DOI:10.1371/journal.pone.0109118 |
Zhang X Y, Wang G H, Xu X Y, Nong X H, Wang J, Amin M, Qi S H. 2016. Exploring fungal diversity in deep-sea sediments from Okinawa Trough using high-throughput Illumina sequencing. Deep Sea Research Part I: Oceanographic Research Papers, 116: 99-105.
DOI:10.1016/j.dsr.2016.08.004 |
Zhang X Y, Zhang Y, Xu X Y, Qi S H. 2013. Diverse deep-sea fungi from the South China Sea and their antimicrobial activity. Curr. Microbiol., 67(5): 525-530.
DOI:10.1007/s00284-013-0394-6 |
 2021, Vol. 39
2021, Vol. 39


